class: center, middle, inverse, title-slide .title[ # Data visualization basics ] .subtitle[ ## – examples in {ggplot2} ] .author[ ### <em>Julia Romanowska</em> ] .date[ ### April 20, 2023 ] --- class: inverse, left, middle ## <svg aria-hidden="true" role="img" viewBox="0 0 640 512" style="height:1em;width:1.25em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:currentColor;overflow:visible;position:relative;"><path d="M64 96c0-35.3 28.7-64 64-64H512c35.3 0 64 28.7 64 64V352H512V96H128V352H64V96zM0 403.2C0 392.6 8.6 384 19.2 384H620.8c10.6 0 19.2 8.6 19.2 19.2c0 42.4-34.4 76.8-76.8 76.8H76.8C34.4 480 0 445.6 0 403.2zM281 209l-31 31 31 31c9.4 9.4 9.4 24.6 0 33.9s-24.6 9.4-33.9 0l-48-48c-9.4-9.4-9.4-24.6 0-33.9l48-48c9.4-9.4 24.6-9.4 33.9 0s9.4 24.6 0 33.9zM393 175l48 48c9.4 9.4 9.4 24.6 0 33.9l-48 48c-9.4 9.4-24.6 9.4-33.9 0s-9.4-24.6 0-33.9l31-31-31-31c-9.4-9.4-9.4-24.6 0-33.9s24.6-9.4 33.9 0z"/></svg> {ggplot2} basics --- ### A plot in {ggplot2} consists of several layers: -- 1. data 2. aesthetics (`aes`) ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-aesthetics)) 3. `geom`s ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-geoms)) 4. `scale`s ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-scales)) 5. `theme` ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-themes)) ??? 1. data - strict format: tidy data! - one row per datapoint - all grouping must be included in the data 2. aesthetics (`aes`) ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-aesthetics)) - how to map the data to the graph? - which column is *x*, *y*, ... - which column provides grouping (based on the grouping, one can either connect points into lines, split a graph into several facets, or color differently each group) 3. `geom`s ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-geoms)) - how to visualize the data? - points (`geom_point`), lines (`geom_line`), bars (`geom_bar`), etc. - `geom`s are connected to `stat`s that conduct any necessary pre-processing of data (e.g., `geom_histogram` would first calculate the number of observations in each bin through `stat_bin`) 4. `scale`s ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-scales)) - any type of data representation on the plot - coordinates: `scale_x_continuous`, `scale_x_discrete`, `scale_x_date`, etc. - colors: `scale_color_manual`, `scale_colour_brewer`, `scale_fill_continuous`, etc. 5. `theme` ([original documentation](https://ggplot2.tidyverse.org/reference/index.html#section-themes)) - visual aspects - size and types of fonts - positioning of axes, legends, etc. Each layer is added to the previous one with a `+` sign. The result can be saved to an object and added upon later. --- class: inverse, left, middle ## Example plot with {ggplot2} --- ## Data <div style="float: right; position: fixed; right: 10px; top: 10px;"> <img style="float: right; position: relative; width: 120px" src="libs/figs/palmer_penguins_logo.png" alt="palmer penguins R package logo"/><br> </div> [palmerpenguins](https://allisonhorst.github.io/palmerpenguins/) <br> ``` # A tibble: 344 × 8 species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 7 Adelie Torgersen 38.9 17.8 181 3625 8 Adelie Torgersen 39.2 19.6 195 4675 9 Adelie Torgersen 34.1 18.1 193 3475 10 Adelie Torgersen 42 20.2 190 4250 # ℹ 334 more rows # ℹ 2 more variables: sex <fct>, year <int> ``` ??? I will be using this simple dataset with morphological data of penguins living on a certain island. -- > **IMPORTANT** start with tidy data! **each row is an observation, each column is a variable** --- count: false .panel1-penguins_species_simple-auto[ ```r *ggplot(data = penguins) ``` ] .panel2-penguins_species_simple-auto[ <!-- --> ] --- count: false .panel1-penguins_species_simple-auto[ ```r ggplot(data = penguins) + * aes( * x = bill_length_mm, * y = bill_depth_mm * ) ``` ] .panel2-penguins_species_simple-auto[ <!-- --> ] --- count: false .panel1-penguins_species_simple-auto[ ```r ggplot(data = penguins) + aes( x = bill_length_mm, y = bill_depth_mm ) + * geom_point() ``` ] .panel2-penguins_species_simple-auto[ <!-- --> ] --- count: false .panel1-penguins_species_simple-auto[ ```r ggplot(data = penguins) + aes( x = bill_length_mm, y = bill_depth_mm ) + geom_point() + * geom_smooth(method = "lm") ``` ] .panel2-penguins_species_simple-auto[ <!-- --> ] <style> .panel1-penguins_species_simple-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_species_simple-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_species_simple-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> --- count: false .panel1-penguins_species-auto[ ```r *ggplot(data = penguins) ``` ] .panel2-penguins_species-auto[ <!-- --> ] --- count: false .panel1-penguins_species-auto[ ```r ggplot(data = penguins) + * aes( * x = bill_length_mm, * y = bill_depth_mm * ) ``` ] .panel2-penguins_species-auto[ <!-- --> ] --- count: false .panel1-penguins_species-auto[ ```r ggplot(data = penguins) + aes( x = bill_length_mm, y = bill_depth_mm ) + * geom_point( * aes(color = species, * shape = species), * size = 2 * ) ``` ] .panel2-penguins_species-auto[ 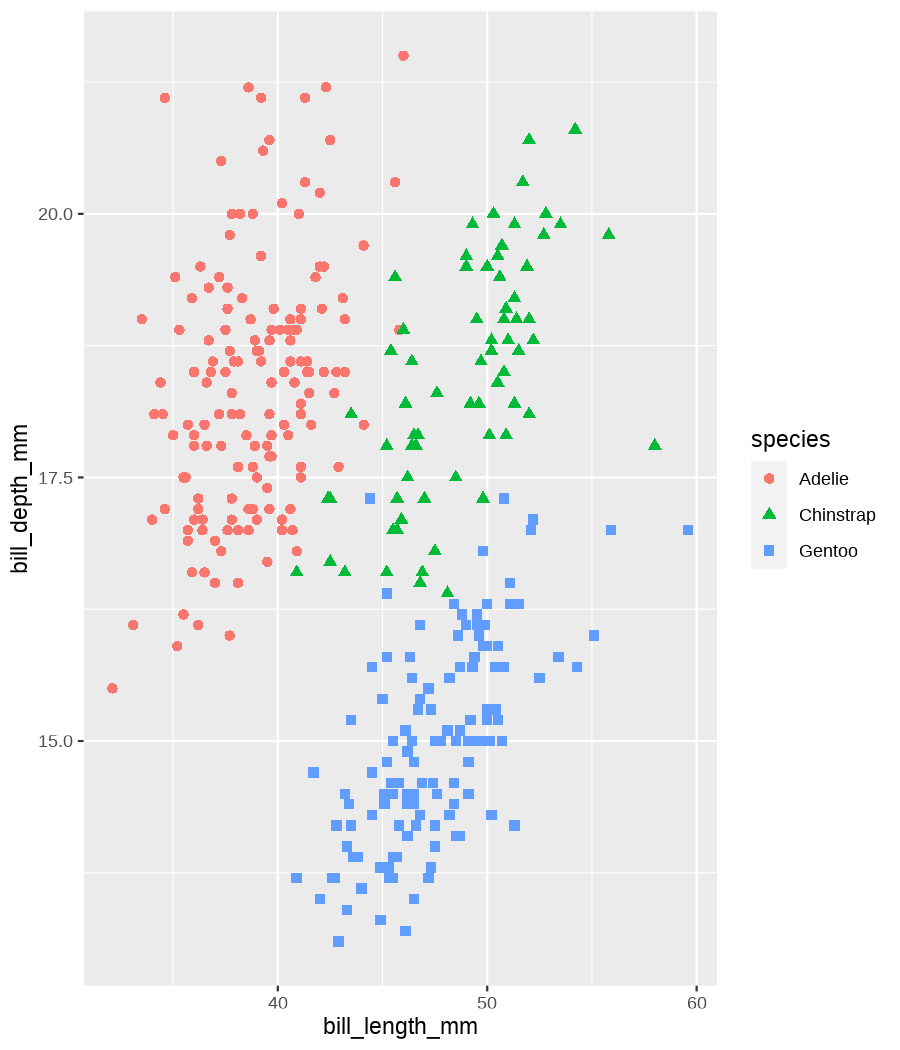<!-- --> ] --- count: false .panel1-penguins_species-auto[ ```r ggplot(data = penguins) + aes( x = bill_length_mm, y = bill_depth_mm ) + geom_point( aes(color = species, shape = species), size = 2 ) + * geom_smooth( * method = "lm", * se = FALSE, * aes(color = species) * ) ``` ] .panel2-penguins_species-auto[ 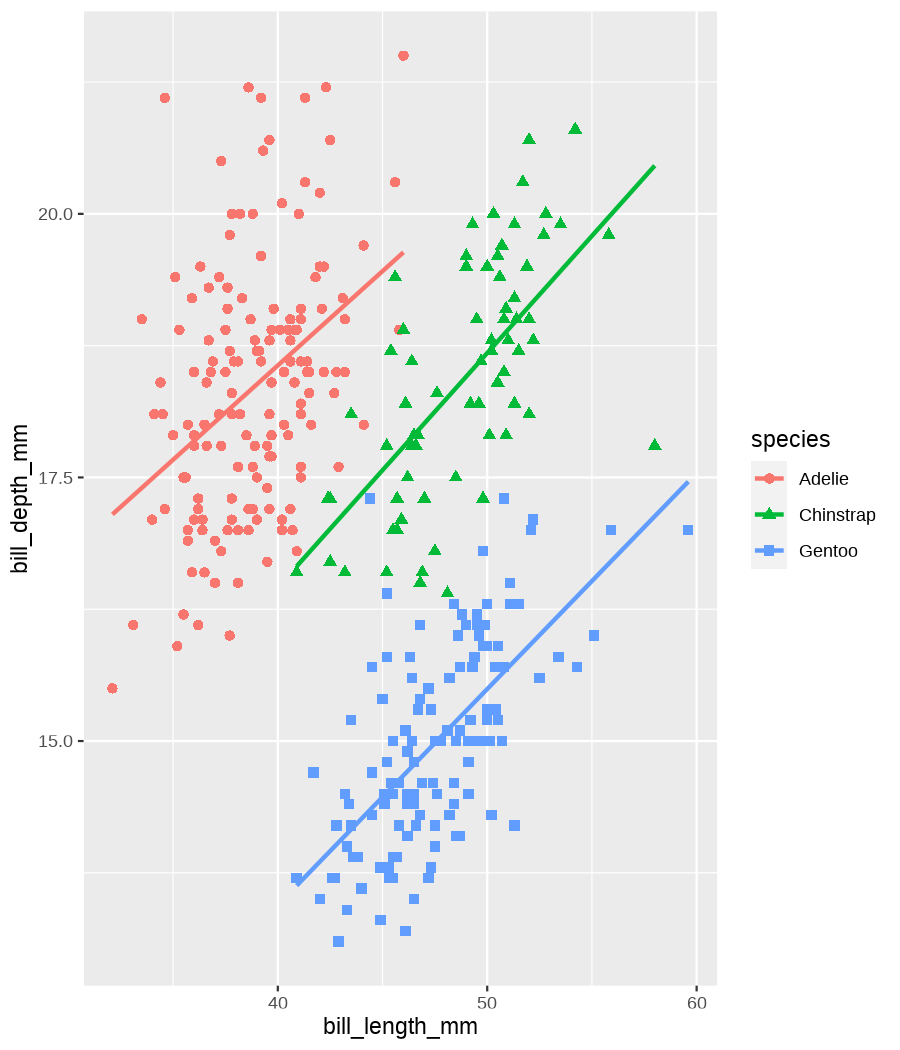<!-- --> ] --- count: false .panel1-penguins_species-auto[ ```r ggplot(data = penguins) + aes( x = bill_length_mm, y = bill_depth_mm ) + geom_point( aes(color = species, shape = species), size = 2 ) + geom_smooth( method = "lm", se = FALSE, aes(color = species) ) *penguins_default <- last_plot() ``` ] .panel2-penguins_species-auto[ 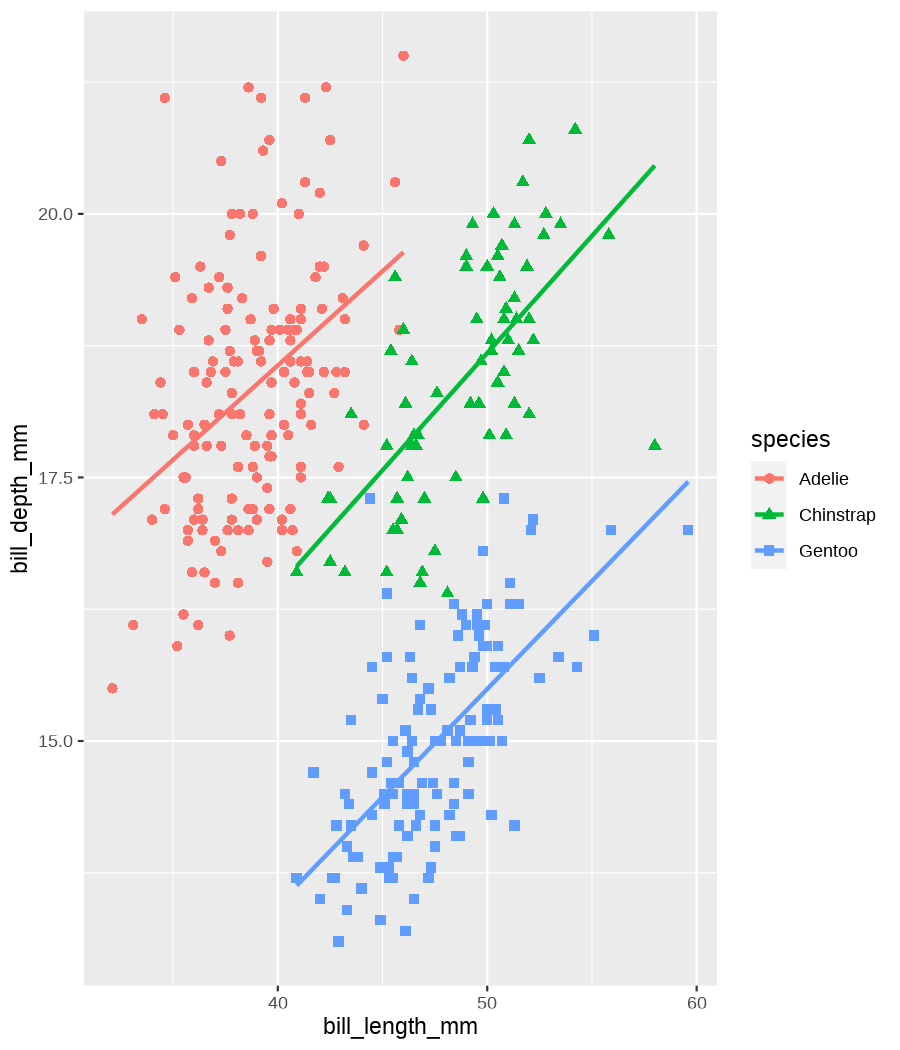<!-- --> ] <style> .panel1-penguins_species-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_species-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_species-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> ??? Basics of ggplot2: - layers - 'aesthetics' + data + geom --- class: inverse, left, middle ## _...back to theory..._ --- ## Bar chart --- count: false .panel1-penguins_barchart-auto[ ```r *ggplot(penguins) ``` ] .panel2-penguins_barchart-auto[ <!-- --> ] --- count: false .panel1-penguins_barchart-auto[ ```r ggplot(penguins) + * geom_bar(aes(island)) ``` ] .panel2-penguins_barchart-auto[ 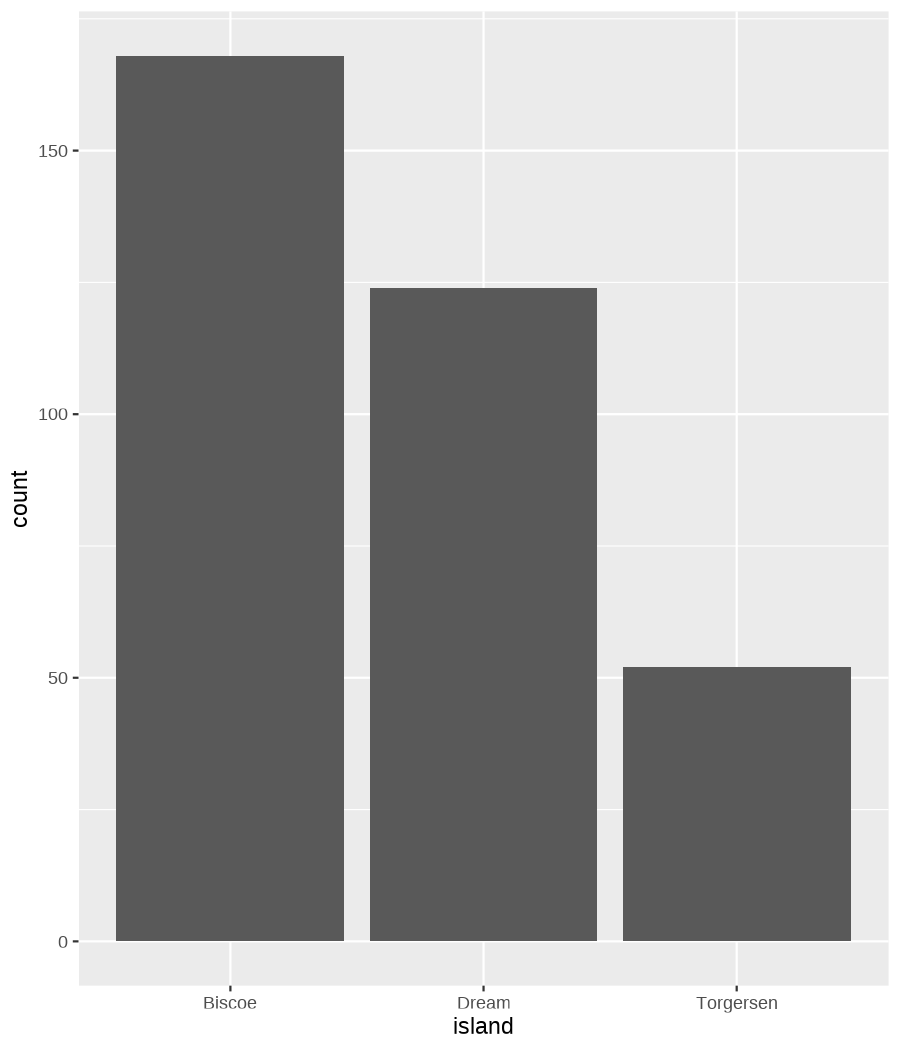<!-- --> ] <style> .panel1-penguins_barchart-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_barchart-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_barchart-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> --- ## Line chart --- count: false .panel1-penguins_linechart-auto[ ```r *penguins ``` ] .panel2-penguins_linechart-auto[ ``` # A tibble: 344 × 8 species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 7 Adelie Torgersen 38.9 17.8 181 3625 8 Adelie Torgersen 39.2 19.6 195 4675 9 Adelie Torgersen 34.1 18.1 193 3475 10 Adelie Torgersen 42 20.2 190 4250 # ℹ 334 more rows # ℹ 2 more variables: sex <fct>, year <int> ``` ] --- count: false .panel1-penguins_linechart-auto[ ```r penguins %>% * count(year, sex) ``` ] .panel2-penguins_linechart-auto[ ``` # A tibble: 9 × 3 year sex n <int> <fct> <int> 1 2007 female 51 2 2007 male 52 3 2007 <NA> 7 4 2008 female 56 5 2008 male 57 6 2008 <NA> 1 7 2009 female 58 8 2009 male 59 9 2009 <NA> 3 ``` ] --- count: false .panel1-penguins_linechart-auto[ ```r penguins %>% count(year, sex) %>% * ggplot(aes(x = year, y = n)) ``` ] .panel2-penguins_linechart-auto[ <!-- --> ] --- count: false .panel1-penguins_linechart-auto[ ```r penguins %>% count(year, sex) %>% ggplot(aes(x = year, y = n)) + * geom_line(aes(color = sex)) ``` ] .panel2-penguins_linechart-auto[ 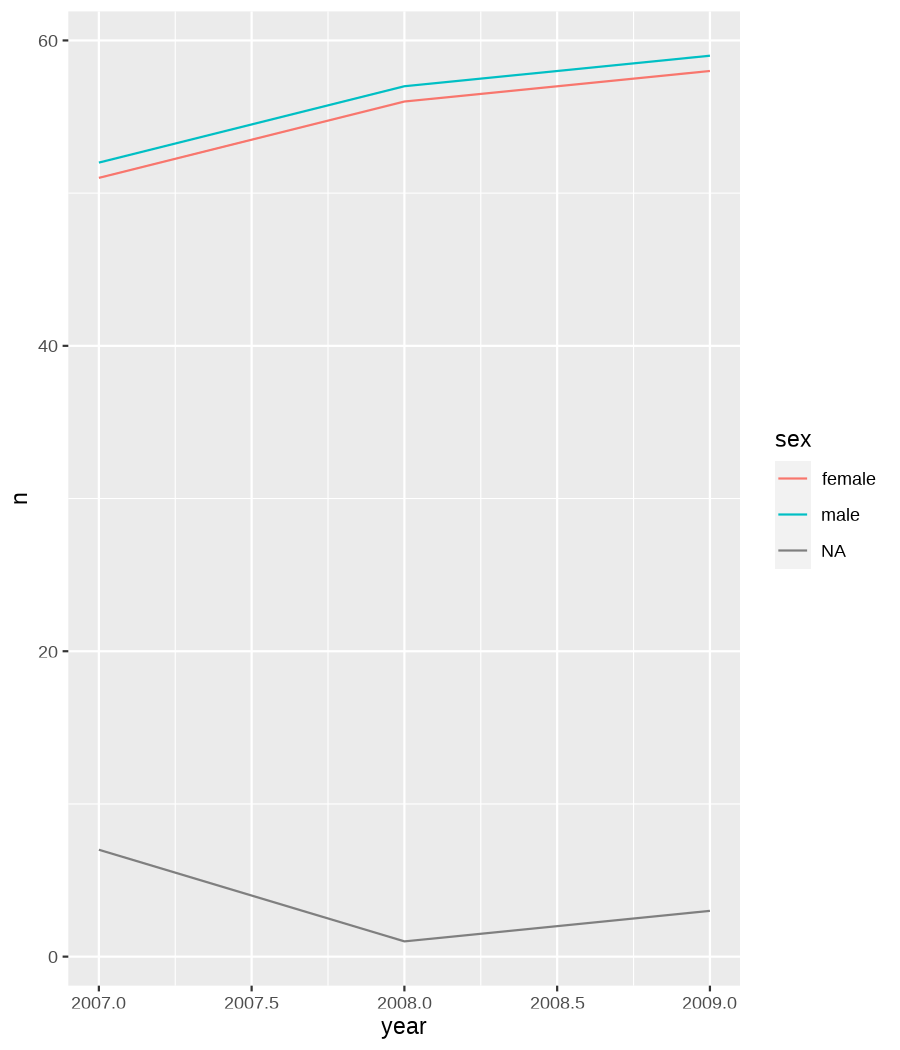<!-- --> ] <style> .panel1-penguins_linechart-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_linechart-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_linechart-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> --- ## Heatmap --- count: false .panel1-penguins_heatmap-auto[ ```r *penguins ``` ] .panel2-penguins_heatmap-auto[ ``` # A tibble: 344 × 8 species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 7 Adelie Torgersen 38.9 17.8 181 3625 8 Adelie Torgersen 39.2 19.6 195 4675 9 Adelie Torgersen 34.1 18.1 193 3475 10 Adelie Torgersen 42 20.2 190 4250 # ℹ 334 more rows # ℹ 2 more variables: sex <fct>, year <int> ``` ] --- count: false .panel1-penguins_heatmap-auto[ ```r penguins %>% * count(species, island) ``` ] .panel2-penguins_heatmap-auto[ ``` # A tibble: 5 × 3 species island n <fct> <fct> <int> 1 Adelie Biscoe 44 2 Adelie Dream 56 3 Adelie Torgersen 52 4 Chinstrap Dream 68 5 Gentoo Biscoe 124 ``` ] --- count: false .panel1-penguins_heatmap-auto[ ```r penguins %>% count(species, island) %>% *ggplot(aes(x = species, y = island)) ``` ] .panel2-penguins_heatmap-auto[ <!-- --> ] --- count: false .panel1-penguins_heatmap-auto[ ```r penguins %>% count(species, island) %>% ggplot(aes(x = species, y = island)) + * geom_tile(aes(fill = n)) ``` ] .panel2-penguins_heatmap-auto[ 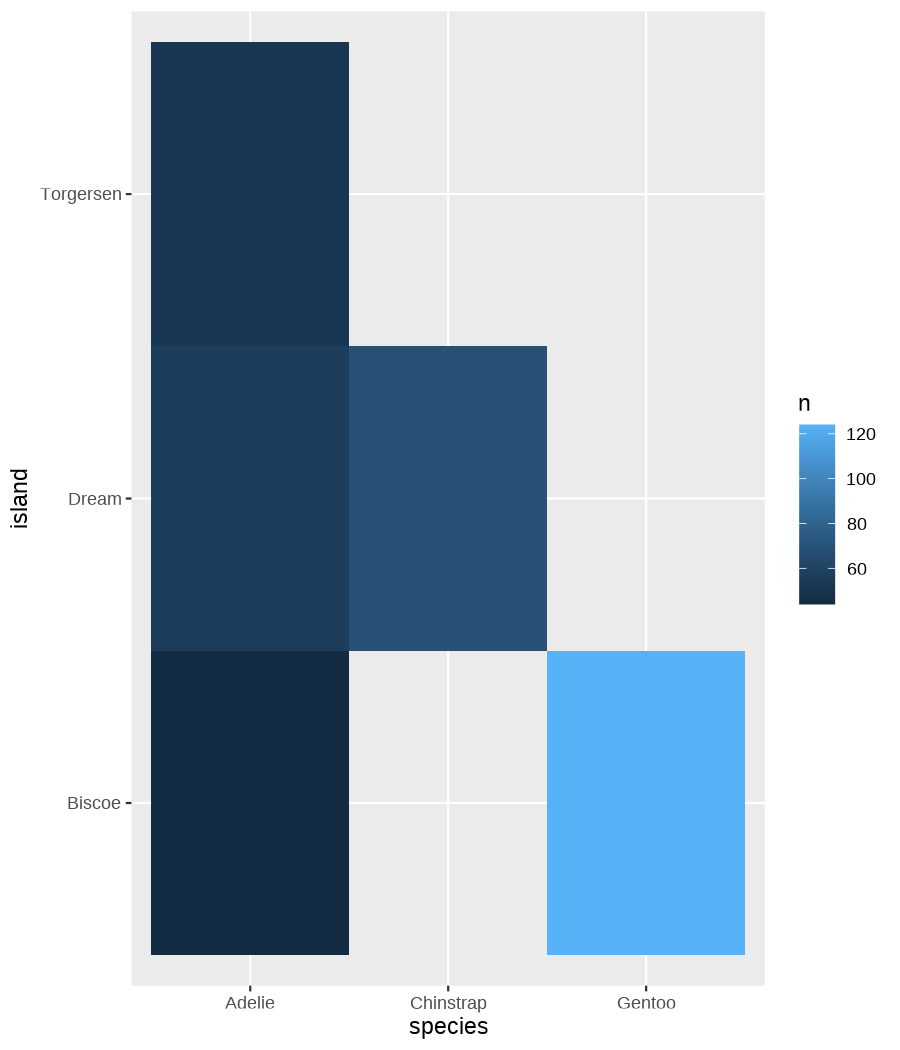<!-- --> ] <style> .panel1-penguins_heatmap-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_heatmap-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_heatmap-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> --- ## Parallel coordinates ### [{ggbump} package](https://github.com/davidsjoberg/ggbump) --- count: false .panel1-penguins_parallel-auto[ ```r *penguins ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 344 × 8 species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 7 Adelie Torgersen 38.9 17.8 181 3625 8 Adelie Torgersen 39.2 19.6 195 4675 9 Adelie Torgersen 34.1 18.1 193 3475 10 Adelie Torgersen 42 20.2 190 4250 # ℹ 334 more rows # ℹ 2 more variables: sex <fct>, year <int> ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% * group_by(species) ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 344 × 8 # Groups: species [3] species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 7 Adelie Torgersen 38.9 17.8 181 3625 8 Adelie Torgersen 39.2 19.6 195 4675 9 Adelie Torgersen 34.1 18.1 193 3475 10 Adelie Torgersen 42 20.2 190 4250 # ℹ 334 more rows # ℹ 2 more variables: sex <fct>, year <int> ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% * summarise( * across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) * ) ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 3 × 5 species bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <dbl> <dbl> <dbl> <dbl> 1 Adelie 38.8 18.3 190. 3701. 2 Chinstrap 48.8 18.4 196. 3733. 3 Gentoo 47.5 15.0 217. 5076. ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% summarise( across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) ) %>% * pivot_longer( * bill_length_mm:body_mass_g, * names_to = "measurement", * values_to = "value" * ) ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 12 × 3 species measurement value <fct> <chr> <dbl> 1 Adelie bill_length_mm 38.8 2 Adelie bill_depth_mm 18.3 3 Adelie flipper_length_mm 190. 4 Adelie body_mass_g 3701. 5 Chinstrap bill_length_mm 48.8 6 Chinstrap bill_depth_mm 18.4 7 Chinstrap flipper_length_mm 196. 8 Chinstrap body_mass_g 3733. 9 Gentoo bill_length_mm 47.5 10 Gentoo bill_depth_mm 15.0 11 Gentoo flipper_length_mm 217. 12 Gentoo body_mass_g 5076. ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% summarise( across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) ) %>% pivot_longer( bill_length_mm:body_mass_g, names_to = "measurement", values_to = "value" ) %>% * group_by(measurement) ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 12 × 3 # Groups: measurement [4] species measurement value <fct> <chr> <dbl> 1 Adelie bill_length_mm 38.8 2 Adelie bill_depth_mm 18.3 3 Adelie flipper_length_mm 190. 4 Adelie body_mass_g 3701. 5 Chinstrap bill_length_mm 48.8 6 Chinstrap bill_depth_mm 18.4 7 Chinstrap flipper_length_mm 196. 8 Chinstrap body_mass_g 3733. 9 Gentoo bill_length_mm 47.5 10 Gentoo bill_depth_mm 15.0 11 Gentoo flipper_length_mm 217. 12 Gentoo body_mass_g 5076. ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% summarise( across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) ) %>% pivot_longer( bill_length_mm:body_mass_g, names_to = "measurement", values_to = "value" ) %>% group_by(measurement) %>% * mutate( * rank = rank(value) * ) ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 12 × 4 # Groups: measurement [4] species measurement value rank <fct> <chr> <dbl> <dbl> 1 Adelie bill_length_mm 38.8 1 2 Adelie bill_depth_mm 18.3 2 3 Adelie flipper_length_mm 190. 1 4 Adelie body_mass_g 3701. 1 5 Chinstrap bill_length_mm 48.8 3 6 Chinstrap bill_depth_mm 18.4 3 7 Chinstrap flipper_length_mm 196. 2 8 Chinstrap body_mass_g 3733. 2 9 Gentoo bill_length_mm 47.5 2 10 Gentoo bill_depth_mm 15.0 1 11 Gentoo flipper_length_mm 217. 3 12 Gentoo body_mass_g 5076. 3 ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% summarise( across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) ) %>% pivot_longer( bill_length_mm:body_mass_g, names_to = "measurement", values_to = "value" ) %>% group_by(measurement) %>% mutate( rank = rank(value) ) %>% * ungroup() ``` ] .panel2-penguins_parallel-auto[ ``` # A tibble: 12 × 4 species measurement value rank <fct> <chr> <dbl> <dbl> 1 Adelie bill_length_mm 38.8 1 2 Adelie bill_depth_mm 18.3 2 3 Adelie flipper_length_mm 190. 1 4 Adelie body_mass_g 3701. 1 5 Chinstrap bill_length_mm 48.8 3 6 Chinstrap bill_depth_mm 18.4 3 7 Chinstrap flipper_length_mm 196. 2 8 Chinstrap body_mass_g 3733. 2 9 Gentoo bill_length_mm 47.5 2 10 Gentoo bill_depth_mm 15.0 1 11 Gentoo flipper_length_mm 217. 3 12 Gentoo body_mass_g 5076. 3 ``` ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% summarise( across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) ) %>% pivot_longer( bill_length_mm:body_mass_g, names_to = "measurement", values_to = "value" ) %>% group_by(measurement) %>% mutate( rank = rank(value) ) %>% ungroup() %>% * ggplot( * aes(x = measurement, y = rank) * ) ``` ] .panel2-penguins_parallel-auto[ <!-- --> ] --- count: false .panel1-penguins_parallel-auto[ ```r penguins %>% group_by(species) %>% summarise( across(bill_length_mm:body_mass_g, ~ mean(.x, na.rm = TRUE)) ) %>% pivot_longer( bill_length_mm:body_mass_g, names_to = "measurement", values_to = "value" ) %>% group_by(measurement) %>% mutate( rank = rank(value) ) %>% ungroup() %>% ggplot( aes(x = measurement, y = rank) ) + * geom_bump(aes(color = species, group = species)) ``` ] .panel2-penguins_parallel-auto[ 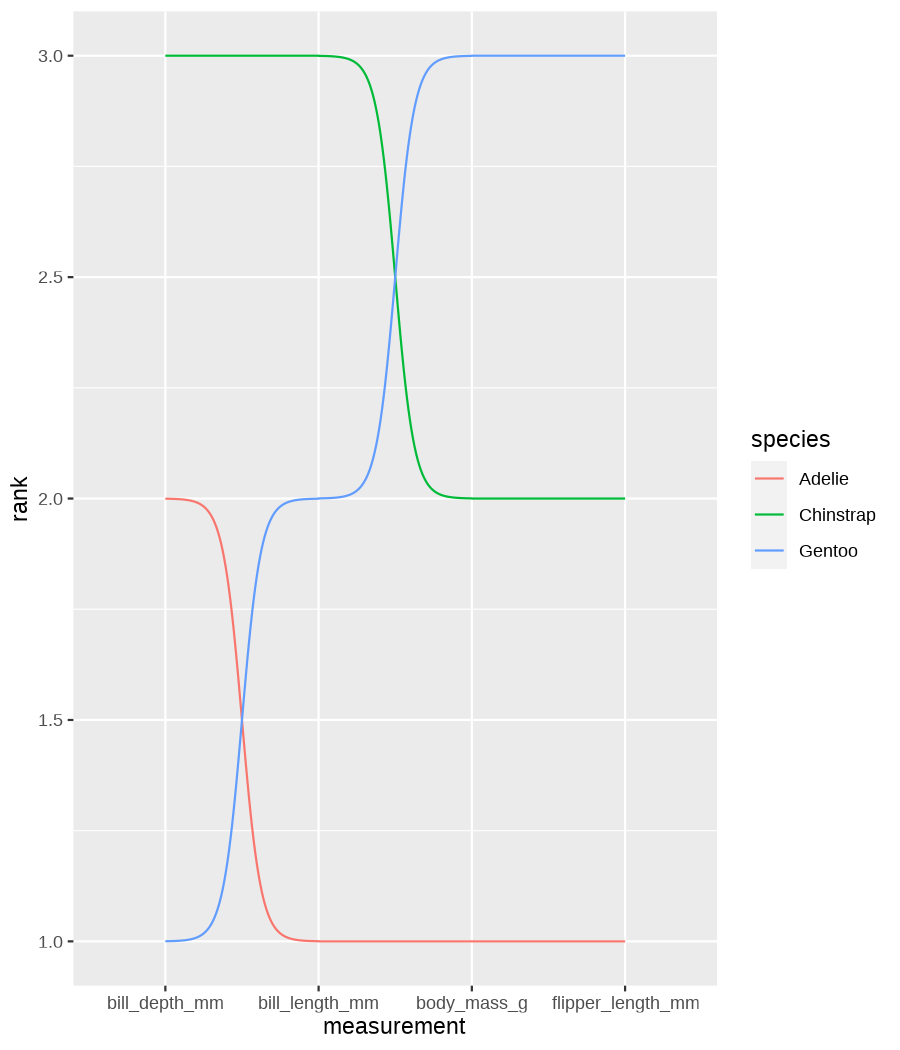<!-- --> ] <style> .panel1-penguins_parallel-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_parallel-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_parallel-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> --- class: left, middle ## ...many other geoms and extensions: https://exts.ggplot2.tidyverse.org/gallery/ --- class: inverse, left, middle ## _...back to theory..._ --- count: false .panel1-penguins_species2-auto[ ```r *penguins_default ``` ] .panel2-penguins_species2-auto[ 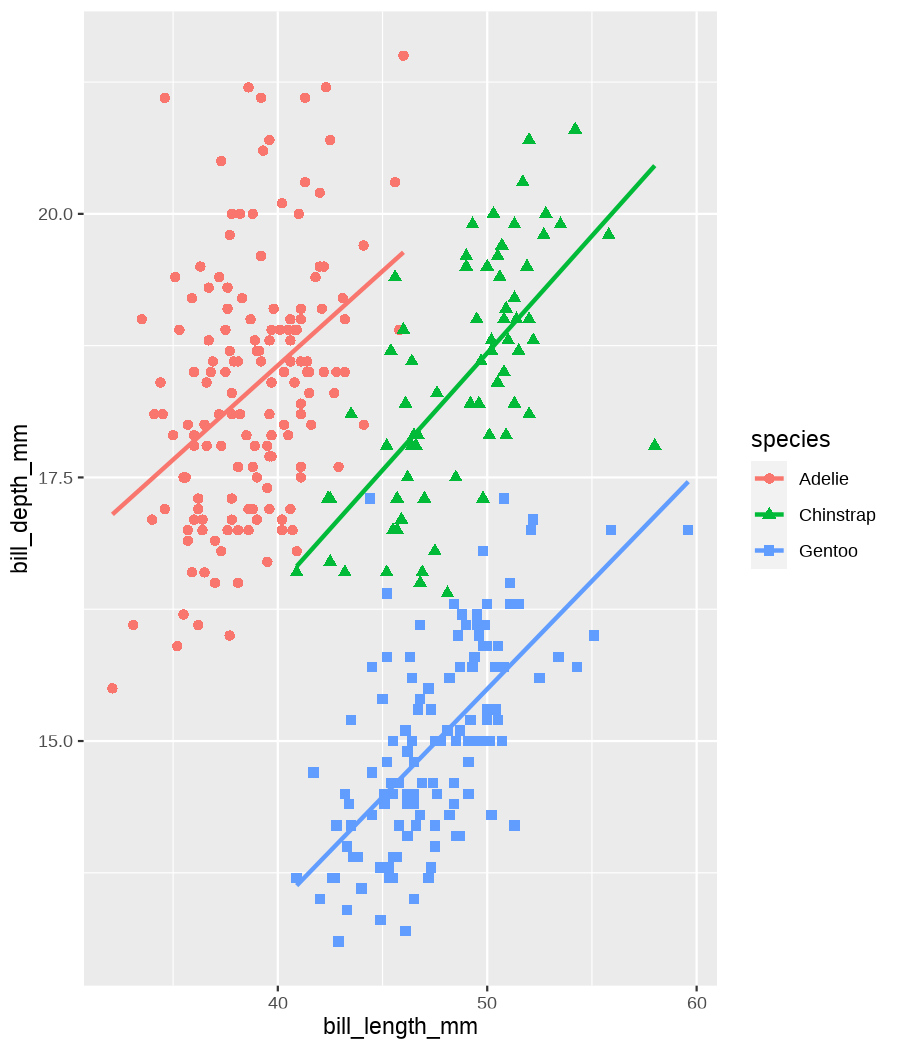<!-- --> ] --- count: false .panel1-penguins_species2-auto[ ```r penguins_default + * facet_grid(rows = vars(species)) ``` ] .panel2-penguins_species2-auto[ 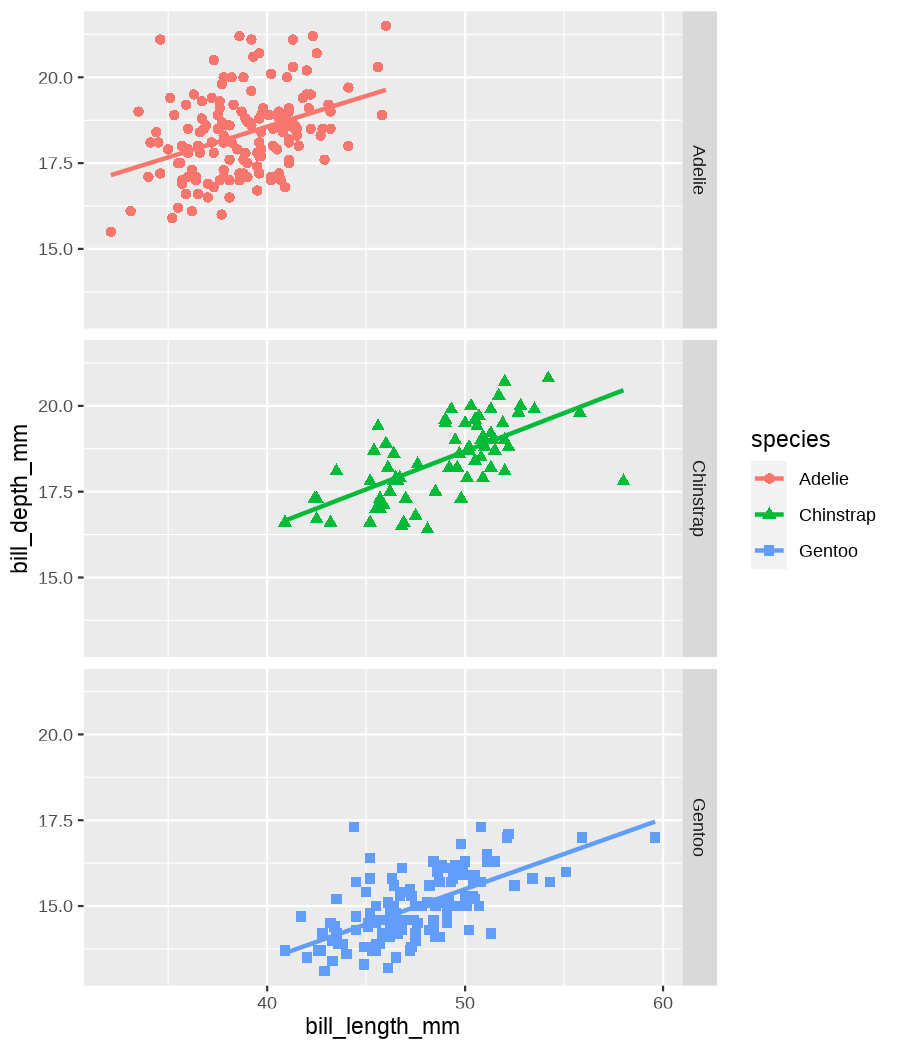<!-- --> ] --- count: false .panel1-penguins_species2-auto[ ```r penguins_default + facet_grid(rows = vars(species)) *penguins_facet <- last_plot() ``` ] .panel2-penguins_species2-auto[ 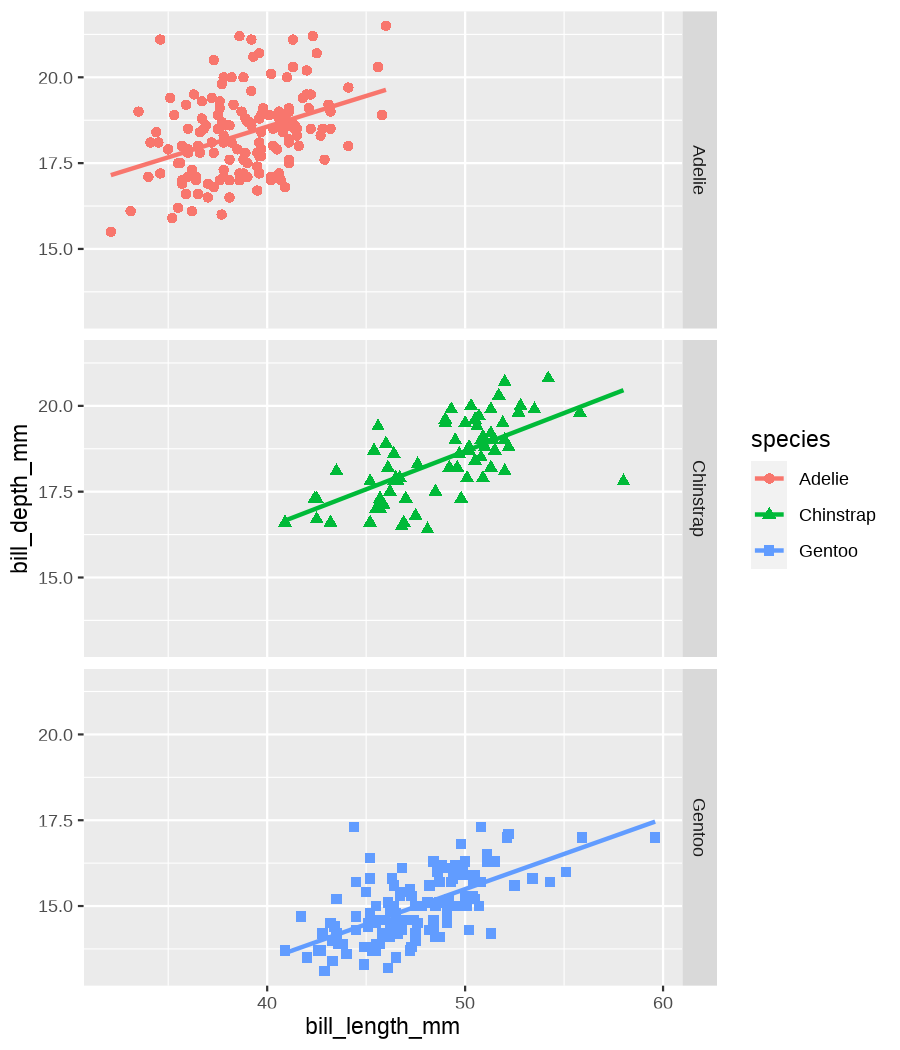<!-- --> ] <style> .panel1-penguins_species2-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_species2-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_species2-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> --- [{ggiraph} package](https://davidgohel.github.io/ggiraph/) .pull-left[ ```r penguins_facet_interactive <- penguins_facet + geom_point_interactive( aes( color = species, shape = species, tooltip = paste0(sex, ", living on ", island) ) ) girafe(ggobj = penguins_facet_interactive) ``` ] .pull-right[ <div class="girafe html-widget html-fill-item-overflow-hidden html-fill-item" id="htmlwidget-1e53680b445e0a399415" style="width:900px;height:1050px;"></div> <script type="application/json" data-for="htmlwidget-1e53680b445e0a399415">{"x":{"html":"<?xml version=\"1.0\" encoding=\"UTF-8\"?>\n<svg xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink' class='ggiraph-svg' role='img' id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65' viewBox='0 0 432 504'>\n <defs id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_defs'>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c1'>\n <rect x='0' y='0' width='432' height='504'/>\n <\/clipPath>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c2'>\n <rect x='43.04' y='5.48' width='279.43' height='151.95'/>\n <\/clipPath>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c3'>\n <rect x='43.04' y='162.91' width='279.43' height='151.95'/>\n <\/clipPath>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c4'>\n <rect x='43.04' y='320.34' width='279.43' height='151.95'/>\n <\/clipPath>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c5'>\n <rect x='322.47' y='5.48' width='17.01' height='151.95'/>\n <\/clipPath>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c6'>\n <rect x='322.47' y='162.91' width='17.01' height='151.95'/>\n <\/clipPath>\n <clipPath id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c7'>\n <rect x='322.47' y='320.34' width='17.01' height='151.95'/>\n <\/clipPath>\n <\/defs>\n <g id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_rootg' class='ggiraph-svg-rootg'>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c1)'>\n <rect x='0' y='0' width='432' height='504' fill='#FFFFFF' fill-opacity='1' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.75' stroke-linejoin='round' stroke-linecap='round' class='ggiraph-svg-bg'/>\n <rect x='0' y='0' width='432' height='504' fill='#FFFFFF' fill-opacity='1' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='round'/>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c2)'>\n <rect x='43.04' y='5.48' width='279.43' height='151.95' fill='#EBEBEB' fill-opacity='1' stroke='none'/>\n <polyline points='43.04,139.84 322.47,139.84' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,98.72 322.47,98.72' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,57.61 322.47,57.61' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,16.50 322.47,16.50' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='82.53,157.43 82.53,5.48' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='174.90,157.43 174.90,5.48' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='267.27,157.43 267.27,5.48' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,119.28 322.47,119.28' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,78.17 322.47,78.17' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,37.05 322.47,37.05' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='128.71,157.43 128.71,5.48' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='221.09,157.43 221.09,5.48' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='313.46,157.43 313.46,5.48' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <circle cx='120.4' cy='58.43' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='124.1' cy='79.81' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='131.49' cy='69.94' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='98.23' cy='48.57' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='122.25' cy='27.19' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='118.55' cy='73.23' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='121.32' cy='43.63' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='74.21' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='147.19' cy='33.76' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='108.39' cy='84.74' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='108.39' cy='81.46' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='76.52' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='115.78' cy='17.32' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='78.83' cy='18.96' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='97.31' cy='73.23' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='116.71' cy='53.5' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='151.81' cy='25.54' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='76.99' cy='63.37' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='184.14' cy='12.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='108.39' cy='65.01' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='107.47' cy='58.43' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='90.84' cy='50.21' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='112.09' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='117.63' cy='83.1' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='85.3' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='134.26' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='133.33' cy='71.59' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='109.32' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='133.33' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='124.1' cy='91.32' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='102.85' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='124.1' cy='73.23' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='137.03' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='95.46' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='121.32' cy='18.96' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='117.63' cy='37.05' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='149.04' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='106.54' cy='48.57' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='126.87' cy='51.85' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='96.38' cy='69.94' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='136.1' cy='63.37' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='91.76' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='166.59' cy='41.99' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='101' cy='88.03' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.02' cy='56.79' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='53.5' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='105.62' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='91.76' cy='71.59' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='149.96' cy='17.32' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.02' cy='74.88' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='129.64' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='82.53' cy='71.59' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='147.19' cy='45.28' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='77.91' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='141.65' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='119.48' cy='78.17' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='134.26' cy='56.79' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='96.38' cy='92.97' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='106.54' cy='51.85' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='88.99' cy='88.03' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='140.72' cy='18.96' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='106.54' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='66.66' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='95.46' cy='84.74' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='143.49' cy='69.94' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='87.15' cy='99.55' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='51.85' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='90.84' cy='92.97' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='145.34' cy='46.92' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='68.67' cy='53.5' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.94' cy='63.37' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.02' cy='83.1' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='182.29' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='87.15' cy='78.17' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='154.58' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='137.03' cy='89.68' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='102.85' cy='46.92' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='93.61' cy='101.19' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='148.11' cy='51.85' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='78.83' cy='83.1' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='155.5' cy='76.52' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='98.23' cy='56.79' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='83.45' cy='46.92' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='103.77' cy='73.23' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='140.72' cy='32.12' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='94.54' cy='45.28' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='100.08' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='113.01' cy='50.21' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='118.55' cy='56.79' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='88.99' cy='69.94' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='73.29' cy='84.74' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.02' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='93.61' cy='81.46' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='136.1' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='111.16' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='131.49' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='64.98' cy='101.19' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='158.27' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='82.53' cy='71.59' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='137.95' cy='37.05' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='107.47' cy='102.83' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='108.39' cy='37.05' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='109.32' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.94' cy='55.14' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='115.78' cy='83.1' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='112.09' cy='37.05' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='111.16' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='158.27' cy='53.5' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='111.16' cy='94.61' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='180.44' cy='32.12' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.94' cy='74.88' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='149.04' cy='45.28' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.02' cy='25.54' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='153.66' cy='65.01' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='115.78' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='103.77' cy='28.83' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='88.99' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='93.61' cy='83.1' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='107.47' cy='40.34' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='130.56' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='141.65' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='84.37' cy='104.48' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='134.26' cy='53.5' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='117.63' cy='76.52' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='142.57' cy='65.01' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='119.48' cy='84.74' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='166.59' cy='69.94' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='114.86' cy='71.59' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='157.35' cy='50.21' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='99.15' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='105.62' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='111.16' cy='76.52' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='138.88' cy='78.17' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='88.07' cy='78.17' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='130.56' cy='35.41' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='101' cy='94.61' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='125.94' cy='71.59' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='130.56' cy='84.74' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='134.26' cy='83.1' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='55.74' cy='111.06' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='135.18' cy='86.39' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='103.77' cy='89.68' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='119.48' cy='58.43' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='121.32' cy='60.08' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='97.31' cy='63.37' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='91.76' cy='73.23' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='108.39' cy='68.3' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='91.76' cy='84.74' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <circle cx='142.57' cy='61.72' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <polyline points='55.74,83.93 57.36,83.41 58.99,82.89 60.62,82.37 62.24,81.86 63.87,81.34 65.49,80.82 67.12,80.30 68.74,79.79 70.37,79.27 71.99,78.75 73.62,78.24 75.24,77.72 76.87,77.20 78.49,76.68 80.12,76.17 81.74,75.65 83.37,75.13 84.99,74.61 86.62,74.10 88.25,73.58 89.87,73.06 91.50,72.54 93.12,72.03 94.75,71.51 96.37,70.99 98.00,70.47 99.62,69.96 101.25,69.44 102.87,68.92 104.50,68.40 106.12,67.89 107.75,67.37 109.37,66.85 111.00,66.33 112.62,65.82 114.25,65.30 115.88,64.78 117.50,64.26 119.13,63.75 120.75,63.23 122.38,62.71 124.00,62.19 125.63,61.68 127.25,61.16 128.88,60.64 130.50,60.12 132.13,59.61 133.75,59.09 135.38,58.57 137.00,58.05 138.63,57.54 140.26,57.02 141.88,56.50 143.51,55.98 145.13,55.47 146.76,54.95 148.38,54.43 150.01,53.91 151.63,53.40 153.26,52.88 154.88,52.36 156.51,51.84 158.13,51.33 159.76,50.81 161.38,50.29 163.01,49.78 164.63,49.26 166.26,48.74 167.89,48.22 169.51,47.71 171.14,47.19 172.76,46.67 174.39,46.15 176.01,45.64 177.64,45.12 179.26,44.60 180.89,44.08 182.51,43.57 184.14,43.05' fill='none' stroke='#F8766D' stroke-opacity='1' stroke-width='2.13' stroke-linejoin='round' stroke-linecap='butt'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e1' cx='120.4' cy='58.43' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e2' cx='124.1' cy='79.81' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e3' cx='131.49' cy='69.94' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e4' cx='98.23' cy='48.57' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e5' cx='122.25' cy='27.19' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e6' cx='118.55' cy='73.23' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e7' cx='121.32' cy='43.63' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e8' cx='74.21' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='NA, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e9' cx='147.19' cy='33.76' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='NA, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e10' cx='108.39' cy='84.74' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='NA, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e11' cx='108.39' cy='81.46' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='NA, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e12' cx='138.88' cy='76.52' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e13' cx='115.78' cy='17.32' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e14' cx='78.83' cy='18.96' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e15' cx='97.31' cy='73.23' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e16' cx='116.71' cy='53.5' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e17' cx='151.81' cy='25.54' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e18' cx='76.99' cy='63.37' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e19' cx='184.14' cy='12.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e20' cx='108.39' cy='65.01' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e21' cx='107.47' cy='58.43' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e22' cx='90.84' cy='50.21' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e23' cx='112.09' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e24' cx='117.63' cy='83.1' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e25' cx='85.3' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e26' cx='134.26' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e27' cx='133.33' cy='71.59' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e28' cx='109.32' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e29' cx='133.33' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e30' cx='124.1' cy='91.32' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e31' cx='102.85' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e32' cx='124.1' cy='73.23' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e33' cx='137.03' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e34' cx='95.46' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e35' cx='121.32' cy='18.96' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e36' cx='117.63' cy='37.05' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e37' cx='149.04' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e38' cx='106.54' cy='48.57' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e39' cx='126.87' cy='51.85' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e40' cx='96.38' cy='69.94' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e41' cx='136.1' cy='63.37' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e42' cx='91.76' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e43' cx='166.59' cy='41.99' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e44' cx='101' cy='88.03' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e45' cx='125.02' cy='56.79' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e46' cx='138.88' cy='53.5' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e47' cx='105.62' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='NA, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e48' cx='91.76' cy='71.59' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e49' cx='149.96' cy='17.32' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e50' cx='125.02' cy='74.88' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e51' cx='129.64' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e52' cx='82.53' cy='71.59' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e53' cx='147.19' cy='45.28' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e54' cx='77.91' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e55' cx='141.65' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e56' cx='119.48' cy='78.17' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e57' cx='134.26' cy='56.79' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e58' cx='96.38' cy='92.97' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e59' cx='106.54' cy='51.85' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e60' cx='88.99' cy='88.03' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e61' cx='140.72' cy='18.96' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e62' cx='106.54' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e63' cx='138.88' cy='66.66' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e64' cx='95.46' cy='84.74' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e65' cx='143.49' cy='69.94' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e66' cx='87.15' cy='99.55' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e67' cx='138.88' cy='51.85' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e68' cx='90.84' cy='92.97' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e69' cx='145.34' cy='46.92' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e70' cx='68.67' cy='53.5' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e71' cx='125.94' cy='63.37' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e72' cx='125.02' cy='83.1' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e73' cx='182.29' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e74' cx='87.15' cy='78.17' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e75' cx='154.58' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e76' cx='137.03' cy='89.68' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e77' cx='102.85' cy='46.92' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e78' cx='93.61' cy='101.19' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e79' cx='148.11' cy='51.85' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e80' cx='78.83' cy='83.1' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e81' cx='155.5' cy='76.52' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e82' cx='98.23' cy='56.79' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e83' cx='83.45' cy='46.92' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e84' cx='103.77' cy='73.23' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e85' cx='140.72' cy='32.12' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e86' cx='94.54' cy='45.28' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e87' cx='100.08' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e88' cx='113.01' cy='50.21' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e89' cx='118.55' cy='56.79' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e90' cx='88.99' cy='69.94' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e91' cx='138.88' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e92' cx='73.29' cy='84.74' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e93' cx='125.02' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e94' cx='93.61' cy='81.46' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e95' cx='136.1' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e96' cx='111.16' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e97' cx='131.49' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e98' cx='64.98' cy='101.19' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e99' cx='158.27' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e100' cx='82.53' cy='71.59' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e101' cx='137.95' cy='37.05' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e102' cx='107.47' cy='102.83' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e103' cx='108.39' cy='37.05' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e104' cx='109.32' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e105' cx='125.94' cy='55.14' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e106' cx='115.78' cy='83.1' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e107' cx='112.09' cy='37.05' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e108' cx='111.16' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e109' cx='158.27' cy='53.5' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e110' cx='111.16' cy='94.61' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e111' cx='180.44' cy='32.12' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e112' cx='125.94' cy='74.88' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e113' cx='149.04' cy='45.28' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e114' cx='125.02' cy='25.54' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e115' cx='153.66' cy='65.01' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e116' cx='115.78' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e117' cx='103.77' cy='28.83' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e118' cx='88.99' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e119' cx='138.88' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e120' cx='93.61' cy='83.1' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e121' cx='107.47' cy='40.34' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e122' cx='130.56' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e123' cx='141.65' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e124' cx='84.37' cy='104.48' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e125' cx='134.26' cy='53.5' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e126' cx='117.63' cy='76.52' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e127' cx='142.57' cy='65.01' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e128' cx='119.48' cy='84.74' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e129' cx='166.59' cy='69.94' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e130' cx='114.86' cy='71.59' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e131' cx='157.35' cy='50.21' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Torgersen'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e132' cx='99.15' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e133' cx='105.62' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e134' cx='111.16' cy='76.52' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e135' cx='138.88' cy='78.17' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e136' cx='88.07' cy='78.17' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e137' cx='130.56' cy='35.41' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e138' cx='101' cy='94.61' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e139' cx='125.94' cy='71.59' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e140' cx='130.56' cy='84.74' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e141' cx='134.26' cy='83.1' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e142' cx='55.74' cy='111.06' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e143' cx='135.18' cy='86.39' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e144' cx='103.77' cy='89.68' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e145' cx='119.48' cy='58.43' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e146' cx='121.32' cy='60.08' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e147' cx='97.31' cy='63.37' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e148' cx='91.76' cy='73.23' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e149' cx='108.39' cy='68.3' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e150' cx='91.76' cy='84.74' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <circle id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e151' cx='142.57' cy='61.72' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c3)'>\n <rect x='43.04' y='162.91' width='279.43' height='151.95' fill='#EBEBEB' fill-opacity='1' stroke='none'/>\n <polyline points='43.04,297.27 322.47,297.27' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,256.15 322.47,256.15' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,215.04 322.47,215.04' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,173.93 322.47,173.93' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='82.53,314.86 82.53,162.91' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='174.90,314.86 174.90,162.91' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='267.27,314.86 267.27,162.91' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,276.71 322.47,276.71' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,235.60 322.47,235.60' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,194.49 322.47,194.49' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='128.71,314.86 128.71,162.91' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='221.09,314.86 221.09,162.91' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='313.46,314.86 313.46,162.91' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polygon points='188.76,225.15 192.11,230.95 185.41,230.95' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='221.09,198.84 224.44,204.64 217.74,204.64' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='233.10,203.77 236.45,209.58 229.74,209.58' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='178.60,211.99 181.95,217.80 175.24,217.80' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='246.03,193.90 249.38,199.71 242.68,199.71' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='176.75,226.80 180.10,232.60 173.40,232.60' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='185.06,220.22 188.41,226.02 181.71,226.02' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='233.10,220.22 236.45,226.02 229.74,226.02' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='184.14,208.71 187.49,214.51 180.79,214.51' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='233.10,192.26 236.45,198.06 229.74,198.06' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='189.68,226.80 193.03,232.60 186.33,232.60' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='236.79,185.68 240.14,191.49 233.44,191.49' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='193.38,235.02 196.73,240.82 190.02,240.82' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='239.56,221.86 242.91,227.67 236.21,227.67' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='183.21,238.31 186.57,244.11 179.86,244.11' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='225.71,197.19 229.06,203.00 222.36,203.00' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='223.86,190.62 227.21,196.42 220.51,196.42' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='294.99,226.80 298.34,232.60 291.64,232.60' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='187.83,213.64 191.18,219.44 184.48,219.44' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='213.70,220.22 217.05,226.02 210.35,226.02' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='150.88,235.02 154.23,240.82 147.53,240.82' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='207.23,231.73 210.58,237.53 203.88,237.53' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='158.27,246.53 161.62,252.33 154.92,252.33' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='226.63,200.48 229.98,206.29 223.28,206.29' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='190.60,225.15 193.96,230.95 187.25,230.95' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='239.56,207.06 242.91,212.87 236.21,212.87' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='225.71,216.93 229.06,222.73 222.36,222.73' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='216.47,207.06 219.82,212.87 213.12,212.87' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='187.83,226.80 191.18,232.60 184.48,232.60' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='246.95,190.62 250.30,196.42 243.60,196.42' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='137.03,246.53 140.38,252.33 133.68,252.33' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='259.88,177.46 263.24,183.26 256.53,183.26' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='151.81,244.88 155.16,250.69 148.46,250.69' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='230.32,210.35 233.68,216.15 226.97,216.15' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='218.32,213.64 221.67,219.44 214.97,219.44' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='197.99,243.24 201.35,249.04 194.64,249.04' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='198.92,218.57 202.27,224.38 195.57,224.38' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='239.56,179.10 242.91,184.91 236.21,184.91' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='192.45,246.53 195.80,252.33 189.10,252.33' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='253.42,192.26 256.77,198.06 250.07,198.06' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='211.85,198.84 215.20,204.64 208.50,204.64' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='185.99,231.73 189.34,237.53 182.63,237.53' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='229.40,205.42 232.75,211.22 226.05,211.22' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='179.52,239.95 182.87,245.76 176.17,245.76' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='229.40,225.15 232.75,230.95 226.05,230.95' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='228.48,215.28 231.83,221.09 225.13,221.09' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='222.01,225.15 225.36,230.95 218.66,230.95' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='211.85,197.19 215.20,203.00 208.50,203.00' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='234.94,211.99 238.29,217.80 231.59,217.80' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='219.24,235.02 222.59,240.82 215.89,240.82' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='203.54,249.82 206.89,255.62 200.19,255.62' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='234.02,207.06 237.37,212.87 230.67,212.87' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='181.37,235.02 184.72,240.82 178.02,240.82' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='227.55,195.55 230.90,201.35 224.20,201.35' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='151.81,235.02 155.16,240.82 148.46,240.82' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='241.41,210.35 244.76,216.15 238.06,216.15' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='176.75,246.53 180.10,252.33 173.40,252.33' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='214.62,192.26 217.97,198.06 211.27,198.06' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='222.94,210.35 226.29,216.15 219.58,216.15' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='180.44,200.48 183.79,206.29 177.09,206.29' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='238.64,198.84 241.99,204.64 235.29,204.64' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='191.53,248.17 194.88,253.98 188.18,253.98' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='181.37,239.95 184.72,245.76 178.02,245.76' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='274.66,193.90 278.02,199.71 271.31,199.71' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='161.04,221.86 164.40,227.67 157.69,227.67' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='217.39,220.22 220.74,226.02 214.04,226.02' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='228.48,207.06 231.83,212.87 225.13,212.87' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polygon points='222.94,211.99 226.29,217.80 219.58,217.80' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <polyline points='137.03,249.45 139.03,248.66 141.03,247.87 143.03,247.08 145.03,246.29 147.03,245.50 149.02,244.71 151.02,243.91 153.02,243.12 155.02,242.33 157.02,241.54 159.02,240.75 161.02,239.96 163.02,239.17 165.02,238.38 167.02,237.59 169.02,236.80 171.02,236.00 173.02,235.21 175.02,234.42 177.02,233.63 179.02,232.84 181.02,232.05 183.02,231.26 185.02,230.47 187.01,229.68 189.01,228.89 191.01,228.09 193.01,227.30 195.01,226.51 197.01,225.72 199.01,224.93 201.01,224.14 203.01,223.35 205.01,222.56 207.01,221.77 209.01,220.98 211.01,220.18 213.01,219.39 215.01,218.60 217.01,217.81 219.01,217.02 221.01,216.23 223.01,215.44 225.00,214.65 227.00,213.86 229.00,213.07 231.00,212.27 233.00,211.48 235.00,210.69 237.00,209.90 239.00,209.11 241.00,208.32 243.00,207.53 245.00,206.74 247.00,205.95 249.00,205.16 251.00,204.37 253.00,203.57 255.00,202.78 257.00,201.99 259.00,201.20 261.00,200.41 262.99,199.62 264.99,198.83 266.99,198.04 268.99,197.25 270.99,196.46 272.99,195.66 274.99,194.87 276.99,194.08 278.99,193.29 280.99,192.50 282.99,191.71 284.99,190.92 286.99,190.13 288.99,189.34 290.99,188.55 292.99,187.75 294.99,186.96' fill='none' stroke='#00BA38' stroke-opacity='1' stroke-width='2.13' stroke-linejoin='round' stroke-linecap='butt'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e152' points='188.76,225.98 191.39,230.54 186.12,230.54' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e153' points='221.09,199.67 223.72,204.23 218.45,204.23' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e154' points='233.10,204.60 235.73,209.16 230.46,209.16' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e155' points='178.60,212.82 181.23,217.38 175.96,217.38' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e156' points='246.03,194.73 248.66,199.29 243.40,199.29' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e157' points='176.75,227.62 179.38,232.18 174.12,232.18' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e158' points='185.06,221.05 187.69,225.61 182.43,225.61' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e159' points='233.10,221.05 235.73,225.61 230.46,225.61' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e160' points='184.14,209.54 186.77,214.10 181.51,214.10' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e161' points='233.10,193.09 235.73,197.65 230.46,197.65' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e162' points='189.68,227.62 192.31,232.18 187.05,232.18' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e163' points='236.79,186.51 239.42,191.07 234.16,191.07' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e164' points='193.38,235.85 196.01,240.41 190.74,240.41' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e165' points='239.56,222.69 242.19,227.25 236.93,227.25' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e166' points='183.21,239.14 185.85,243.70 180.58,243.70' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e167' points='225.71,198.02 228.34,202.58 223.07,202.58' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e168' points='223.86,191.45 226.49,196.01 221.23,196.01' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e169' points='294.99,227.62 297.62,232.18 292.35,232.18' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e170' points='187.83,214.47 190.47,219.03 185.20,219.03' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e171' points='213.70,221.05 216.33,225.61 211.07,225.61' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e172' points='150.88,235.85 153.52,240.41 148.25,240.41' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e173' points='207.23,232.56 209.86,237.12 204.60,237.12' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e174' points='158.27,247.36 160.91,251.92 155.64,251.92' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e175' points='226.63,201.31 229.26,205.87 224.00,205.87' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e176' points='190.60,225.98 193.24,230.54 187.97,230.54' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e177' points='239.56,207.89 242.19,212.45 236.93,212.45' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e178' points='225.71,217.76 228.34,222.32 223.07,222.32' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e179' points='216.47,207.89 219.10,212.45 213.84,212.45' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e180' points='187.83,227.62 190.47,232.18 185.20,232.18' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e181' points='246.95,191.45 249.58,196.01 244.32,196.01' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e182' points='137.03,247.36 139.66,251.92 134.40,251.92' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e183' points='259.88,178.29 262.52,182.85 257.25,182.85' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e184' points='151.81,245.71 154.44,250.27 149.17,250.27' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e185' points='230.32,211.18 232.96,215.74 227.69,215.74' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e186' points='218.32,214.47 220.95,219.03 215.68,219.03' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e187' points='197.99,244.07 200.63,248.63 195.36,248.63' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e188' points='198.92,219.40 201.55,223.96 196.29,223.96' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e189' points='239.56,179.93 242.19,184.49 236.93,184.49' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e190' points='192.45,247.36 195.08,251.92 189.82,251.92' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e191' points='253.42,193.09 256.05,197.65 250.79,197.65' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e192' points='211.85,199.67 214.48,204.23 209.22,204.23' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e193' points='185.99,232.56 188.62,237.12 183.35,237.12' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e194' points='229.40,206.25 232.03,210.81 226.77,210.81' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e195' points='179.52,240.78 182.15,245.34 176.89,245.34' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e196' points='229.40,225.98 232.03,230.54 226.77,230.54' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e197' points='228.48,216.11 231.11,220.67 225.84,220.67' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e198' points='222.01,225.98 224.64,230.54 219.38,230.54' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e199' points='211.85,198.02 214.48,202.58 209.22,202.58' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e200' points='234.94,212.82 237.58,217.38 232.31,217.38' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e201' points='219.24,235.85 221.87,240.41 216.61,240.41' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e202' points='203.54,250.65 206.17,255.21 200.90,255.21' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e203' points='234.02,207.89 236.65,212.45 231.39,212.45' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e204' points='181.37,235.85 184.00,240.41 178.73,240.41' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e205' points='227.55,196.38 230.19,200.94 224.92,200.94' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e206' points='151.81,235.85 154.44,240.41 149.17,240.41' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e207' points='241.41,211.18 244.04,215.74 238.78,215.74' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e208' points='176.75,247.36 179.38,251.92 174.12,251.92' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e209' points='214.62,193.09 217.25,197.65 211.99,197.65' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e210' points='222.94,211.18 225.57,215.74 220.30,215.74' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e211' points='180.44,201.31 183.08,205.87 177.81,205.87' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e212' points='238.64,199.67 241.27,204.23 236.01,204.23' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e213' points='191.53,249.00 194.16,253.56 188.90,253.56' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e214' points='181.37,240.78 184.00,245.34 178.73,245.34' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e215' points='274.66,194.73 277.30,199.29 272.03,199.29' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e216' points='161.04,222.69 163.68,227.25 158.41,227.25' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e217' points='217.39,221.05 220.03,225.61 214.76,225.61' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e218' points='228.48,207.89 231.11,212.45 225.84,212.45' fill='#00BA38' fill-opacity='1' stroke='none' title='male, living on Dream'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e219' points='222.94,212.82 225.57,217.38 220.30,217.38' fill='#00BA38' fill-opacity='1' stroke='none' title='female, living on Dream'/>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c4)'>\n <rect x='43.04' y='320.34' width='279.43' height='151.95' fill='#EBEBEB' fill-opacity='1' stroke='none'/>\n <polyline points='43.04,454.70 322.47,454.70' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,413.59 322.47,413.59' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,372.47 322.47,372.47' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,331.36 322.47,331.36' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='82.53,472.29 82.53,320.34' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='174.90,472.29 174.90,320.34' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='267.27,472.29 267.27,320.34' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.53' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,434.14 322.47,434.14' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,393.03 322.47,393.03' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='43.04,351.92 322.47,351.92' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='128.71,472.29 128.71,320.34' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='221.09,472.29 221.09,320.34' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='313.46,472.29 313.46,320.34' fill='none' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polygon points='182.57,466.23 187.55,466.23 187.55,461.26 182.57,461.26' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='218.60,415.25 223.58,415.25 223.58,410.28 218.60,410.28' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='206.59,451.43 211.57,451.43 211.57,446.45 206.59,446.45' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='218.60,433.34 223.58,433.34 223.58,428.37 218.60,428.37' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='196.43,444.85 201.41,444.85 201.41,439.88 196.43,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='186.27,461.30 191.25,461.30 191.25,456.32 186.27,456.32' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='176.11,443.21 181.08,443.21 181.08,438.23 176.11,438.23' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='188.12,431.70 193.09,431.70 193.09,426.72 188.12,426.72' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='156.71,462.94 161.69,462.94 161.69,457.97 156.71,457.97' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='189.04,430.05 194.02,430.05 194.02,425.08 189.04,425.08' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='134.54,458.01 139.52,458.01 139.52,453.03 134.54,453.03' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='209.36,418.54 214.34,418.54 214.34,413.56 209.36,413.56' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='177.03,458.01 182.01,458.01 182.01,453.03 177.03,453.03' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='203.82,443.21 208.80,443.21 208.80,438.23 203.82,438.23' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='179.80,443.21 184.78,443.21 184.78,438.23 179.80,438.23' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='212.13,425.12 217.11,425.12 217.11,420.14 212.13,420.14' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='144.70,461.30 149.68,461.30 149.68,456.32 144.70,456.32' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='211.21,433.34 216.19,433.34 216.19,428.37 211.21,428.37' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='183.50,444.85 188.47,444.85 188.47,439.88 183.50,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='206.59,434.99 211.57,434.99 211.57,430.01 206.59,430.01' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='220.45,448.14 225.42,448.14 225.42,443.17 220.45,443.17' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='173.34,444.85 178.31,444.85 178.31,439.88 173.34,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='186.27,444.85 191.25,444.85 191.25,439.88 186.27,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='184.42,423.47 189.40,423.47 189.40,418.50 184.42,418.50' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='153.01,467.88 157.99,467.88 157.99,462.90 153.01,462.90' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='182.57,434.99 187.55,434.99 187.55,430.01 182.57,430.01' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='167.79,448.14 172.77,448.14 172.77,443.17 167.79,443.17' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='198.28,436.63 203.25,436.63 203.25,431.65 198.28,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='201.97,448.14 206.95,448.14 206.95,443.17 201.97,443.17' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='218.60,431.70 223.58,431.70 223.58,426.72 218.60,426.72' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='193.66,431.70 198.64,431.70 198.64,426.72 193.66,426.72' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='152.09,449.79 157.07,449.79 157.07,444.81 152.09,444.81' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='173.34,444.85 178.31,444.85 178.31,439.88 173.34,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='307.28,403.74 312.25,403.74 312.25,398.76 307.28,398.76' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='210.29,439.92 215.26,439.92 215.26,434.94 210.29,434.94' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='203.82,415.25 208.80,415.25 208.80,410.28 203.82,410.28' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='150.24,458.01 155.22,458.01 155.22,453.03 150.24,453.03' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='166.87,398.81 171.85,398.81 171.85,393.83 166.87,393.83' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='163.18,459.65 168.15,459.65 168.15,454.68 163.18,454.68' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='206.59,425.12 211.57,425.12 211.57,420.14 206.59,420.14' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='151.17,458.01 156.14,458.01 156.14,453.03 151.17,453.03' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='214.90,420.19 219.88,420.19 219.88,415.21 214.90,415.21' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='175.18,458.01 180.16,458.01 180.16,453.03 175.18,453.03' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='214.90,436.63 219.88,436.63 219.88,431.65 214.90,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='223.22,421.83 228.19,421.83 228.19,416.85 223.22,416.85' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='159.48,454.72 164.46,454.72 164.46,449.74 159.48,449.74' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='177.03,454.72 182.01,454.72 182.01,449.74 177.03,449.74' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='223.22,421.83 228.19,421.83 228.19,416.85 223.22,416.85' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='171.49,464.59 176.47,464.59 176.47,459.61 171.49,459.61' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='174.26,423.47 179.24,423.47 179.24,418.50 174.26,418.50' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='187.19,449.79 192.17,449.79 192.17,444.81 187.19,444.81' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='204.74,451.43 209.72,451.43 209.72,446.45 204.74,446.45' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='173.34,446.50 178.31,446.50 178.31,441.52 173.34,441.52' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='219.52,436.63 224.50,436.63 224.50,431.65 219.52,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='186.27,446.50 191.25,446.50 191.25,441.52 186.27,441.52' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='172.41,430.05 177.39,430.05 177.39,425.08 172.41,425.08' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='161.33,454.72 166.30,454.72 166.30,449.74 161.33,449.74' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='177.03,436.63 182.01,436.63 182.01,431.65 177.03,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='155.79,444.85 160.76,444.85 160.76,439.88 155.79,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='222.29,431.70 227.27,431.70 227.27,426.72 222.29,426.72' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='175.18,456.36 180.16,456.36 180.16,451.39 175.18,451.39' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='183.50,438.28 188.47,438.28 188.47,433.30 183.50,433.30' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='178.88,454.72 183.86,454.72 183.86,449.74 178.88,449.74' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='258.32,425.12 263.30,425.12 263.30,420.14 258.32,420.14' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='179.80,449.79 184.78,449.79 184.78,444.81 179.80,444.81' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='216.75,407.03 221.73,407.03 221.73,402.05 216.75,402.05' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='183.50,446.50 188.47,446.50 188.47,441.52 183.50,441.52' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='213.98,416.90 218.96,416.90 218.96,411.92 213.98,411.92' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='158.56,449.79 163.53,449.79 163.53,444.81 158.56,444.81' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='225.07,436.63 230.04,436.63 230.04,431.65 225.07,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='197.35,436.63 202.33,436.63 202.33,431.65 197.35,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='185.34,426.76 190.32,426.76 190.32,421.79 185.34,421.79' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='201.97,426.76 206.95,426.76 206.95,421.79 201.97,421.79' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='186.27,439.92 191.25,439.92 191.25,434.94 186.27,434.94' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='185.34,436.63 190.32,436.63 190.32,431.65 185.34,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='205.67,420.19 210.64,420.19 210.64,415.21 205.67,415.21' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='195.51,449.79 200.48,449.79 200.48,444.81 195.51,444.81' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='228.76,415.25 233.74,415.25 233.74,410.28 228.76,410.28' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='174.26,456.36 179.24,456.36 179.24,451.39 174.26,451.39' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='174.26,413.61 179.24,413.61 179.24,408.63 174.26,408.63' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='210.29,444.85 215.26,444.85 215.26,439.88 210.29,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='241.69,426.76 246.67,426.76 246.67,421.79 241.69,421.79' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='194.58,443.21 199.56,443.21 199.56,438.23 194.58,438.23' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='218.60,421.83 223.58,421.83 223.58,416.85 218.60,416.85' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='171.49,456.36 176.47,456.36 176.47,451.39 171.49,451.39' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='225.99,398.81 230.97,398.81 230.97,393.83 225.99,393.83' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='157.63,446.50 162.61,446.50 162.61,441.52 157.63,441.52' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='230.61,449.79 235.58,449.79 235.58,444.81 230.61,444.81' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='195.51,453.08 200.48,453.08 200.48,448.10 195.51,448.10' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='238.00,403.74 242.97,403.74 242.97,398.76 238.00,398.76' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='195.51,436.63 200.48,436.63 200.48,431.65 195.51,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='238.92,402.10 243.90,402.10 243.90,397.12 238.92,397.12' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='177.03,444.85 182.01,444.85 182.01,439.88 177.03,439.88' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='213.98,418.54 218.96,418.54 218.96,413.56 213.98,413.56' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='167.79,441.56 172.77,441.56 172.77,436.59 167.79,436.59' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='225.99,425.12 230.97,425.12 230.97,420.14 225.99,420.14' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='213.06,423.47 218.03,423.47 218.03,418.50 213.06,418.50' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='189.96,443.21 194.94,443.21 194.94,438.23 189.96,438.23' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='203.82,446.50 208.80,446.50 208.80,441.52 203.82,441.52' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='228.76,411.96 233.74,411.96 233.74,406.99 228.76,406.99' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='204.74,436.63 209.72,436.63 209.72,431.65 204.74,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='273.10,403.74 278.08,403.74 278.08,398.76 273.10,398.76' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='192.73,428.41 197.71,428.41 197.71,423.43 192.73,423.43' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='210.29,436.63 215.26,436.63 215.26,431.65 210.29,431.65' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='193.66,456.36 198.64,456.36 198.64,451.39 193.66,451.39' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='189.04,418.54 194.02,418.54 194.02,413.56 189.04,413.56' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='141.93,441.56 146.91,441.56 146.91,436.59 141.93,436.59' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='250.01,423.47 254.98,423.47 254.98,418.50 250.01,418.50' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='156.71,453.08 161.69,453.08 161.69,448.10 156.71,448.10' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='201.05,434.99 206.02,434.99 206.02,430.01 201.05,430.01' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='223.22,433.34 228.19,433.34 228.19,428.37 223.22,428.37' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='216.75,421.83 221.73,421.83 221.73,416.85 216.75,416.85' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='158.56,433.34 163.53,433.34 163.53,428.37 158.56,428.37' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='232.46,415.25 237.43,415.25 237.43,410.28 232.46,410.28' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='183.50,451.43 188.47,451.43 188.47,446.45 183.50,446.45' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='265.71,420.19 270.69,420.19 270.69,415.21 265.71,415.21' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='167.79,425.12 172.77,425.12 172.77,420.14 167.79,420.14' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='207.51,416.90 212.49,416.90 212.49,411.92 207.51,411.92' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='192.73,458.01 197.71,458.01 197.71,453.03 192.73,453.03' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='189.04,448.14 194.02,448.14 194.02,443.17 189.04,443.17' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='222.29,425.12 227.27,425.12 227.27,420.14 222.29,420.14' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='174.26,439.92 179.24,439.92 179.24,434.94 174.26,434.94' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polygon points='217.68,418.54 222.65,418.54 222.65,413.56 217.68,413.56' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <polyline points='137.03,456.69 139.21,455.89 141.40,455.09 143.59,454.29 145.77,453.50 147.96,452.70 150.15,451.90 152.33,451.10 154.52,450.31 156.71,449.51 158.89,448.71 161.08,447.91 163.27,447.12 165.45,446.32 167.64,445.52 169.83,444.73 172.01,443.93 174.20,443.13 176.39,442.33 178.57,441.54 180.76,440.74 182.95,439.94 185.13,439.14 187.32,438.35 189.51,437.55 191.69,436.75 193.88,435.95 196.06,435.16 198.25,434.36 200.44,433.56 202.62,432.76 204.81,431.97 207.00,431.17 209.18,430.37 211.37,429.57 213.56,428.78 215.74,427.98 217.93,427.18 220.12,426.39 222.30,425.59 224.49,424.79 226.68,423.99 228.86,423.20 231.05,422.40 233.24,421.60 235.42,420.80 237.61,420.01 239.80,419.21 241.98,418.41 244.17,417.61 246.36,416.82 248.54,416.02 250.73,415.22 252.92,414.42 255.10,413.63 257.29,412.83 259.48,412.03 261.66,411.23 263.85,410.44 266.03,409.64 268.22,408.84 270.41,408.05 272.59,407.25 274.78,406.45 276.97,405.65 279.15,404.86 281.34,404.06 283.53,403.26 285.71,402.46 287.90,401.67 290.09,400.87 292.27,400.07 294.46,399.27 296.65,398.48 298.83,397.68 301.02,396.88 303.21,396.08 305.39,395.29 307.58,394.49 309.77,393.69' fill='none' stroke='#619CFF' stroke-opacity='1' stroke-width='2.13' stroke-linejoin='round' stroke-linecap='butt'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e220' points='183.11,465.70 187.02,465.70 187.02,461.79 183.11,461.79' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e221' points='219.13,414.72 223.04,414.72 223.04,410.81 219.13,410.81' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e222' points='207.12,450.90 211.03,450.90 211.03,446.99 207.12,446.99' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e223' points='219.13,432.81 223.04,432.81 223.04,428.90 219.13,428.90' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e224' points='196.96,444.32 200.87,444.32 200.87,440.41 196.96,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e225' points='186.80,460.76 190.71,460.76 190.71,456.86 186.80,456.86' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e226' points='176.64,442.68 180.55,442.68 180.55,438.77 176.64,438.77' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e227' points='188.65,431.16 192.56,431.16 192.56,427.25 188.65,427.25' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e228' points='157.24,462.41 161.15,462.41 161.15,458.50 157.24,458.50' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e229' points='189.57,429.52 193.48,429.52 193.48,425.61 189.57,425.61' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e230' points='135.07,457.48 138.98,457.48 138.98,453.57 135.07,453.57' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e231' points='209.90,418.01 213.81,418.01 213.81,414.10 209.90,414.10' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e232' points='177.56,457.48 181.47,457.48 181.47,453.57 177.56,453.57' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e233' points='204.35,442.68 208.26,442.68 208.26,438.77 204.35,438.77' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e234' points='180.34,442.68 184.25,442.68 184.25,438.77 180.34,438.77' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e235' points='212.67,424.59 216.58,424.59 216.58,420.68 212.67,420.68' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e236' points='145.23,460.76 149.14,460.76 149.14,456.86 145.23,456.86' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e237' points='211.74,432.81 215.65,432.81 215.65,428.90 211.74,428.90' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e238' points='184.03,444.32 187.94,444.32 187.94,440.41 184.03,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e239' points='207.12,434.45 211.03,434.45 211.03,430.54 207.12,430.54' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e240' points='220.98,447.61 224.89,447.61 224.89,443.70 220.98,443.70' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e241' points='173.87,444.32 177.78,444.32 177.78,440.41 173.87,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e242' points='186.80,444.32 190.71,444.32 190.71,440.41 186.80,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e243' points='184.95,422.94 188.86,422.94 188.86,419.03 184.95,419.03' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e244' points='153.55,467.34 157.46,467.34 157.46,463.43 153.55,463.43' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e245' points='183.11,434.45 187.02,434.45 187.02,430.54 183.11,430.54' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e246' points='168.33,447.61 172.24,447.61 172.24,443.70 168.33,443.70' fill='#619CFF' fill-opacity='1' stroke='none' title='NA, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e247' points='198.81,436.10 202.72,436.10 202.72,432.19 198.81,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e248' points='202.51,447.61 206.42,447.61 206.42,443.70 202.51,443.70' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e249' points='219.13,431.16 223.04,431.16 223.04,427.25 219.13,427.25' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e250' points='194.19,431.16 198.10,431.16 198.10,427.25 194.19,427.25' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e251' points='152.62,449.25 156.53,449.25 156.53,445.34 152.62,445.34' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e252' points='173.87,444.32 177.78,444.32 177.78,440.41 173.87,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e253' points='307.81,403.21 311.72,403.21 311.72,399.30 307.81,399.30' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e254' points='210.82,439.39 214.73,439.39 214.73,435.48 210.82,435.48' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e255' points='204.35,414.72 208.26,414.72 208.26,410.81 204.35,410.81' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e256' points='150.78,457.48 154.69,457.48 154.69,453.57 150.78,453.57' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e257' points='167.40,398.27 171.31,398.27 171.31,394.36 167.40,394.36' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e258' points='163.71,459.12 167.62,459.12 167.62,455.21 163.71,455.21' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e259' points='207.12,424.59 211.03,424.59 211.03,420.68 207.12,420.68' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e260' points='151.70,457.48 155.61,457.48 155.61,453.57 151.70,453.57' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e261' points='215.44,419.65 219.35,419.65 219.35,415.74 215.44,415.74' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e262' points='175.72,457.48 179.63,457.48 179.63,453.57 175.72,453.57' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e263' points='215.44,436.10 219.35,436.10 219.35,432.19 215.44,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e264' points='223.75,421.30 227.66,421.30 227.66,417.39 223.75,417.39' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e265' points='160.01,454.19 163.92,454.19 163.92,450.28 160.01,450.28' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e266' points='177.56,454.19 181.47,454.19 181.47,450.28 177.56,450.28' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e267' points='223.75,421.30 227.66,421.30 227.66,417.39 223.75,417.39' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e268' points='172.02,464.05 175.93,464.05 175.93,460.14 172.02,460.14' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e269' points='174.79,422.94 178.70,422.94 178.70,419.03 174.79,419.03' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e270' points='187.73,449.25 191.64,449.25 191.64,445.34 187.73,445.34' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e271' points='205.28,450.90 209.19,450.90 209.19,446.99 205.28,446.99' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e272' points='173.87,445.96 177.78,445.96 177.78,442.05 173.87,442.05' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e273' points='220.06,436.10 223.97,436.10 223.97,432.19 220.06,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e274' points='186.80,445.96 190.71,445.96 190.71,442.05 186.80,442.05' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e275' points='172.95,429.52 176.86,429.52 176.86,425.61 172.95,425.61' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e276' points='161.86,454.19 165.77,454.19 165.77,450.28 161.86,450.28' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e277' points='177.56,436.10 181.47,436.10 181.47,432.19 177.56,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e278' points='156.32,444.32 160.23,444.32 160.23,440.41 156.32,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e279' points='222.83,431.16 226.74,431.16 226.74,427.25 222.83,427.25' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e280' points='175.72,455.83 179.63,455.83 179.63,451.92 175.72,451.92' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e281' points='184.03,437.74 187.94,437.74 187.94,433.83 184.03,433.83' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e282' points='179.41,454.19 183.32,454.19 183.32,450.28 179.41,450.28' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e283' points='258.85,424.59 262.76,424.59 262.76,420.68 258.85,420.68' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e284' points='180.34,449.25 184.25,449.25 184.25,445.34 180.34,445.34' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e285' points='217.29,406.50 221.19,406.50 221.19,402.59 217.29,402.59' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e286' points='184.03,445.96 187.94,445.96 187.94,442.05 184.03,442.05' fill='#619CFF' fill-opacity='1' stroke='none' title='NA, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e287' points='214.51,416.36 218.42,416.36 218.42,412.45 214.51,412.45' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e288' points='159.09,449.25 163.00,449.25 163.00,445.34 159.09,445.34' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e289' points='225.60,436.10 229.51,436.10 229.51,432.19 225.60,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e290' points='197.89,436.10 201.80,436.10 201.80,432.19 197.89,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e291' points='185.88,426.23 189.79,426.23 189.79,422.32 185.88,422.32' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e292' points='202.51,426.23 206.42,426.23 206.42,422.32 202.51,422.32' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e293' points='186.80,439.39 190.71,439.39 190.71,435.48 186.80,435.48' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e294' points='185.88,436.10 189.79,436.10 189.79,432.19 185.88,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e295' points='206.20,419.65 210.11,419.65 210.11,415.74 206.20,415.74' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e296' points='196.04,449.25 199.95,449.25 199.95,445.34 196.04,445.34' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e297' points='229.29,414.72 233.20,414.72 233.20,410.81 229.29,410.81' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e298' points='174.79,455.83 178.70,455.83 178.70,451.92 174.79,451.92' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e299' points='174.79,413.07 178.70,413.07 178.70,409.16 174.79,409.16' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e300' points='210.82,444.32 214.73,444.32 214.73,440.41 210.82,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e301' points='242.23,426.23 246.14,426.23 246.14,422.32 242.23,422.32' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e302' points='195.12,442.68 199.03,442.68 199.03,438.77 195.12,438.77' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e303' points='219.13,421.30 223.04,421.30 223.04,417.39 219.13,417.39' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e304' points='172.02,455.83 175.93,455.83 175.93,451.92 172.02,451.92' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e305' points='226.52,398.27 230.43,398.27 230.43,394.36 226.52,394.36' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e306' points='158.17,445.96 162.08,445.96 162.08,442.05 158.17,442.05' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e307' points='231.14,449.25 235.05,449.25 235.05,445.34 231.14,445.34' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e308' points='196.04,452.54 199.95,452.54 199.95,448.63 196.04,448.63' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e309' points='238.53,403.21 242.44,403.21 242.44,399.30 238.53,399.30' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e310' points='196.04,436.10 199.95,436.10 199.95,432.19 196.04,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e311' points='239.45,401.56 243.36,401.56 243.36,397.65 239.45,397.65' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e312' points='177.56,444.32 181.47,444.32 181.47,440.41 177.56,440.41' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e313' points='214.51,418.01 218.42,418.01 218.42,414.10 214.51,414.10' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e314' points='168.33,441.03 172.24,441.03 172.24,437.12 168.33,437.12' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e315' points='226.52,424.59 230.43,424.59 230.43,420.68 226.52,420.68' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e316' points='213.59,422.94 217.50,422.94 217.50,419.03 213.59,419.03' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e317' points='190.50,442.68 194.41,442.68 194.41,438.77 190.50,438.77' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e318' points='204.35,445.96 208.26,445.96 208.26,442.05 204.35,442.05' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e319' points='229.29,411.43 233.20,411.43 233.20,407.52 229.29,407.52' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e320' points='205.28,436.10 209.19,436.10 209.19,432.19 205.28,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e321' points='273.63,403.21 277.54,403.21 277.54,399.30 273.63,399.30' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e322' points='193.27,427.87 197.18,427.87 197.18,423.97 193.27,423.97' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e323' points='210.82,436.10 214.73,436.10 214.73,432.19 210.82,432.19' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e324' points='194.19,455.83 198.10,455.83 198.10,451.92 194.19,451.92' fill='#619CFF' fill-opacity='1' stroke='none' title='NA, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e325' points='189.57,418.01 193.48,418.01 193.48,414.10 189.57,414.10' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e326' points='142.46,441.03 146.37,441.03 146.37,437.12 142.46,437.12' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e327' points='250.54,422.94 254.45,422.94 254.45,419.03 250.54,419.03' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e328' points='157.24,452.54 161.15,452.54 161.15,448.63 157.24,448.63' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e329' points='201.58,434.45 205.49,434.45 205.49,430.54 201.58,430.54' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e330' points='223.75,432.81 227.66,432.81 227.66,428.90 223.75,428.90' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e331' points='217.29,421.30 221.19,421.30 221.19,417.39 217.29,417.39' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e332' points='159.09,432.81 163.00,432.81 163.00,428.90 159.09,428.90' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e333' points='232.99,414.72 236.90,414.72 236.90,410.81 232.99,410.81' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e334' points='184.03,450.90 187.94,450.90 187.94,446.99 184.03,446.99' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e335' points='266.24,419.65 270.15,419.65 270.15,415.74 266.24,415.74' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e336' points='168.33,424.59 172.24,424.59 172.24,420.68 168.33,420.68' fill='#619CFF' fill-opacity='1' stroke='none' title='NA, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e337' points='208.05,416.36 211.96,416.36 211.96,412.45 208.05,412.45' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e338' points='193.27,457.48 197.18,457.48 197.18,453.57 193.27,453.57' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e339' points='189.57,447.61 193.48,447.61 193.48,443.70 189.57,443.70' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e340' points='222.83,424.59 226.74,424.59 226.74,420.68 222.83,420.68' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e341' points='174.79,439.39 178.70,439.39 178.70,435.48 174.79,435.48' fill='#619CFF' fill-opacity='1' stroke='none' title='female, living on Biscoe'/>\n <polygon id='svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_e342' points='218.21,418.01 222.12,418.01 222.12,414.10 218.21,414.10' fill='#619CFF' fill-opacity='1' stroke='none' title='male, living on Biscoe'/>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c5)'>\n <rect x='322.47' y='5.48' width='17.01' height='151.95' fill='#D9D9D9' fill-opacity='1' stroke='none'/>\n <text transform='translate(327.77,67.87) rotate(90.00)' font-size='6.6pt' font-family='DejaVu Sans' fill='#1A1A1A' fill-opacity='1'>Adelie<\/text>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c6)'>\n <rect x='322.47' y='162.91' width='17.01' height='151.95' fill='#D9D9D9' fill-opacity='1' stroke='none'/>\n <text transform='translate(327.77,217.70) rotate(90.00)' font-size='6.6pt' font-family='DejaVu Sans' fill='#1A1A1A' fill-opacity='1'>Chinstrap<\/text>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c7)'>\n <rect x='322.47' y='320.34' width='17.01' height='151.95' fill='#D9D9D9' fill-opacity='1' stroke='none'/>\n <text transform='translate(327.77,380.31) rotate(90.00)' font-size='6.6pt' font-family='DejaVu Sans' fill='#1A1A1A' fill-opacity='1'>Gentoo<\/text>\n <\/g>\n <g clip-path='url(#svg_d7091ea1_cfe0_40eb_a156_2ef56563df65_c1)'>\n <polyline points='128.71,475.03 128.71,472.29' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='221.09,475.03 221.09,472.29' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='313.46,475.03 313.46,472.29' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <text x='123.12' y='483.64' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>40<\/text>\n <text x='215.49' y='483.64' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>50<\/text>\n <text x='307.87' y='483.64' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>60<\/text>\n <text x='18.53' y='122.49' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>15.0<\/text>\n <text x='18.53' y='81.37' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>17.5<\/text>\n <text x='18.53' y='40.26' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>20.0<\/text>\n <polyline points='40.30,119.28 43.04,119.28' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.30,78.17 43.04,78.17' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.30,37.05 43.04,37.05' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <text x='18.53' y='279.92' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>15.0<\/text>\n <text x='18.53' y='238.81' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>17.5<\/text>\n <text x='18.53' y='197.69' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>20.0<\/text>\n <polyline points='40.30,276.71 43.04,276.71' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.30,235.60 43.04,235.60' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.30,194.49 43.04,194.49' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <text x='18.53' y='437.35' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>15.0<\/text>\n <text x='18.53' y='396.24' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>17.5<\/text>\n <text x='18.53' y='355.12' font-size='6.6pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>20.0<\/text>\n <polyline points='40.30,434.14 43.04,434.14' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.30,393.03 43.04,393.03' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.30,351.92 43.04,351.92' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.07' stroke-linejoin='round' stroke-linecap='butt'/>\n <text x='140.92' y='496.23' font-size='8.25pt' font-family='DejaVu Sans'>bill_length_mm<\/text>\n <text transform='translate(13.50,279.19) rotate(-90.00)' font-size='8.25pt' font-family='DejaVu Sans'>bill_depth_mm<\/text>\n <rect x='350.44' y='199.59' width='76.08' height='78.59' fill='#FFFFFF' fill-opacity='1' stroke='none'/>\n <text x='355.92' y='214.24' font-size='8.25pt' font-family='DejaVu Sans'>species<\/text>\n <rect x='355.92' y='220.86' width='17.28' height='17.28' fill='#F2F2F2' fill-opacity='1' stroke='none'/>\n <circle cx='364.56' cy='229.5' r='1.87pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <line x1='357.64' y1='229.5' x2='371.47' y2='229.5' stroke='#F8766D' stroke-opacity='1' stroke-width='2.13' stroke-linejoin='round' stroke-linecap='butt'/>\n <circle cx='364.56' cy='229.5' r='1.47pt' fill='#F8766D' fill-opacity='1' stroke='none'/>\n <rect x='355.92' y='238.14' width='17.28' height='17.28' fill='#F2F2F2' fill-opacity='1' stroke='none'/>\n <polygon points='364.56,242.91 367.91,248.72 361.21,248.72' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <line x1='357.64' y1='246.78' x2='371.47' y2='246.78' stroke='#00BA38' stroke-opacity='1' stroke-width='2.13' stroke-linejoin='round' stroke-linecap='butt'/>\n <polygon points='364.56,243.74 367.19,248.30 361.92,248.30' fill='#00BA38' fill-opacity='1' stroke='none'/>\n <rect x='355.92' y='255.42' width='17.28' height='17.28' fill='#F2F2F2' fill-opacity='1' stroke='none'/>\n <polygon points='362.07,266.55 367.05,266.55 367.05,261.57 362.07,261.57' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <line x1='357.64' y1='264.06' x2='371.47' y2='264.06' stroke='#619CFF' stroke-opacity='1' stroke-width='2.13' stroke-linejoin='round' stroke-linecap='butt'/>\n <polygon points='362.60,266.02 366.51,266.02 366.51,262.11 362.60,262.11' fill='#619CFF' fill-opacity='1' stroke='none'/>\n <text x='378.68' y='232.71' font-size='6.6pt' font-family='DejaVu Sans'>Adelie<\/text>\n <text x='378.68' y='249.99' font-size='6.6pt' font-family='DejaVu Sans'>Chinstrap<\/text>\n <text x='378.68' y='267.27' font-size='6.6pt' font-family='DejaVu Sans'>Gentoo<\/text>\n <\/g>\n <\/g>\n<\/svg>","js":null,"uid":"svg_d7091ea1_cfe0_40eb_a156_2ef56563df65","ratio":0.857142857142857,"settings":{"tooltip":{"css":".tooltip_SVGID_ { padding:5px;background:black;color:white;border-radius:2px;text-align:left; ; position:absolute;pointer-events:none;z-index:999;}","placement":"doc","opacity":0.9,"offx":10,"offy":10,"use_cursor_pos":true,"use_fill":false,"use_stroke":false,"delay_over":200,"delay_out":500},"hover":{"css":".hover_data_SVGID_ { fill:orange;stroke:black;cursor:pointer; }\ntext.hover_data_SVGID_ { stroke:none;fill:orange; }\ncircle.hover_data_SVGID_ { fill:orange;stroke:black; }\nline.hover_data_SVGID_, polyline.hover_data_SVGID_ { fill:none;stroke:orange; }\nrect.hover_data_SVGID_, polygon.hover_data_SVGID_, path.hover_data_SVGID_ { fill:orange;stroke:none; }\nimage.hover_data_SVGID_ { stroke:orange; }","reactive":true,"nearest_distance":null},"hover_inv":{"css":""},"hover_key":{"css":".hover_key_SVGID_ { fill:orange;stroke:black;cursor:pointer; }\ntext.hover_key_SVGID_ { stroke:none;fill:orange; }\ncircle.hover_key_SVGID_ { fill:orange;stroke:black; }\nline.hover_key_SVGID_, polyline.hover_key_SVGID_ { fill:none;stroke:orange; }\nrect.hover_key_SVGID_, polygon.hover_key_SVGID_, path.hover_key_SVGID_ { fill:orange;stroke:none; }\nimage.hover_key_SVGID_ { stroke:orange; }","reactive":true},"hover_theme":{"css":".hover_theme_SVGID_ { fill:orange;stroke:black;cursor:pointer; }\ntext.hover_theme_SVGID_ { stroke:none;fill:orange; }\ncircle.hover_theme_SVGID_ { fill:orange;stroke:black; }\nline.hover_theme_SVGID_, polyline.hover_theme_SVGID_ { fill:none;stroke:orange; }\nrect.hover_theme_SVGID_, polygon.hover_theme_SVGID_, path.hover_theme_SVGID_ { fill:orange;stroke:none; }\nimage.hover_theme_SVGID_ { stroke:orange; }","reactive":true},"select":{"css":".select_data_SVGID_ { fill:red;stroke:black;cursor:pointer; }\ntext.select_data_SVGID_ { stroke:none;fill:red; }\ncircle.select_data_SVGID_ { fill:red;stroke:black; }\nline.select_data_SVGID_, polyline.select_data_SVGID_ { fill:none;stroke:red; }\nrect.select_data_SVGID_, polygon.select_data_SVGID_, path.select_data_SVGID_ { fill:red;stroke:none; }\nimage.select_data_SVGID_ { stroke:red; }","type":"multiple","only_shiny":true,"selected":[]},"select_inv":{"css":""},"select_key":{"css":".select_key_SVGID_ { fill:red;stroke:black;cursor:pointer; }\ntext.select_key_SVGID_ { stroke:none;fill:red; }\ncircle.select_key_SVGID_ { fill:red;stroke:black; }\nline.select_key_SVGID_, polyline.select_key_SVGID_ { fill:none;stroke:red; }\nrect.select_key_SVGID_, polygon.select_key_SVGID_, path.select_key_SVGID_ { fill:red;stroke:none; }\nimage.select_key_SVGID_ { stroke:red; }","type":"single","only_shiny":true,"selected":[]},"select_theme":{"css":".select_theme_SVGID_ { fill:red;stroke:black;cursor:pointer; }\ntext.select_theme_SVGID_ { stroke:none;fill:red; }\ncircle.select_theme_SVGID_ { fill:red;stroke:black; }\nline.select_theme_SVGID_, polyline.select_theme_SVGID_ { fill:none;stroke:red; }\nrect.select_theme_SVGID_, polygon.select_theme_SVGID_, path.select_theme_SVGID_ { fill:red;stroke:none; }\nimage.select_theme_SVGID_ { stroke:red; }","type":"single","only_shiny":true,"selected":[]},"zoom":{"min":1,"max":1,"duration":300},"toolbar":{"position":"topright","pngname":"diagram","tooltips":null,"hidden":[],"delay_over":200,"delay_out":500},"sizing":{"rescale":true,"width":1}}},"evals":[],"jsHooks":[]}</script> ] --- class: inverse, left, middle ## _...back to theory..._ --- count: false .panel1-penguins_species3-auto[ ```r *penguins_facet ``` ] .panel2-penguins_species3-auto[ 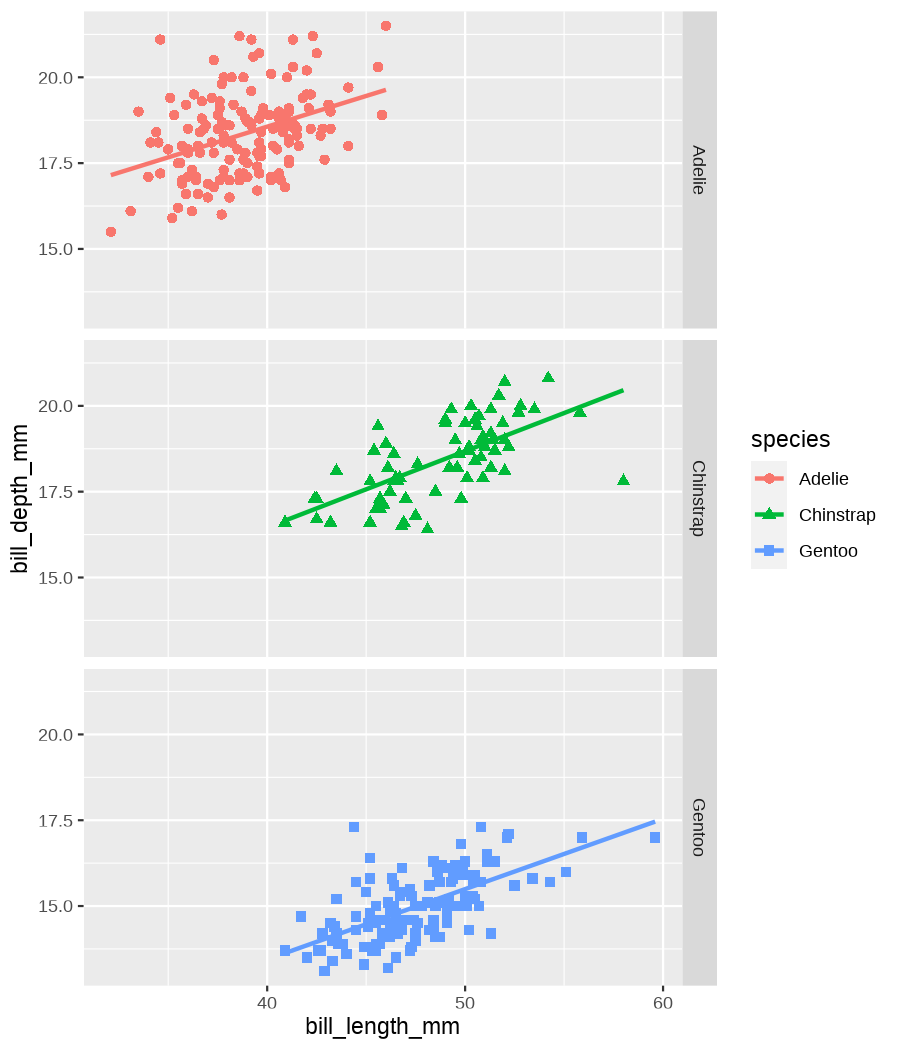<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + * scale_color_manual( * values = c("darkorange", * "darkorchid", * "cyan4") * ) ``` ] .panel2-penguins_species3-auto[ 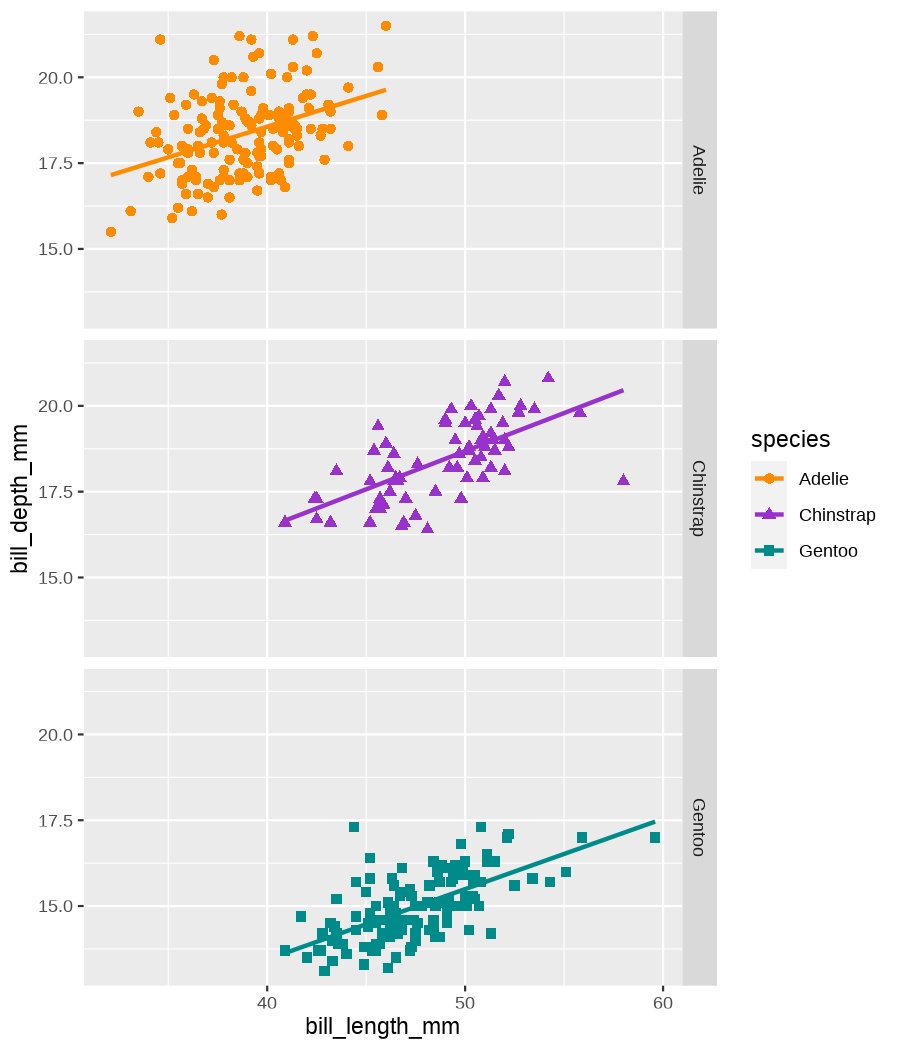<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + * labs(title = * "Penguin species differentation") ``` ] .panel2-penguins_species3-auto[ 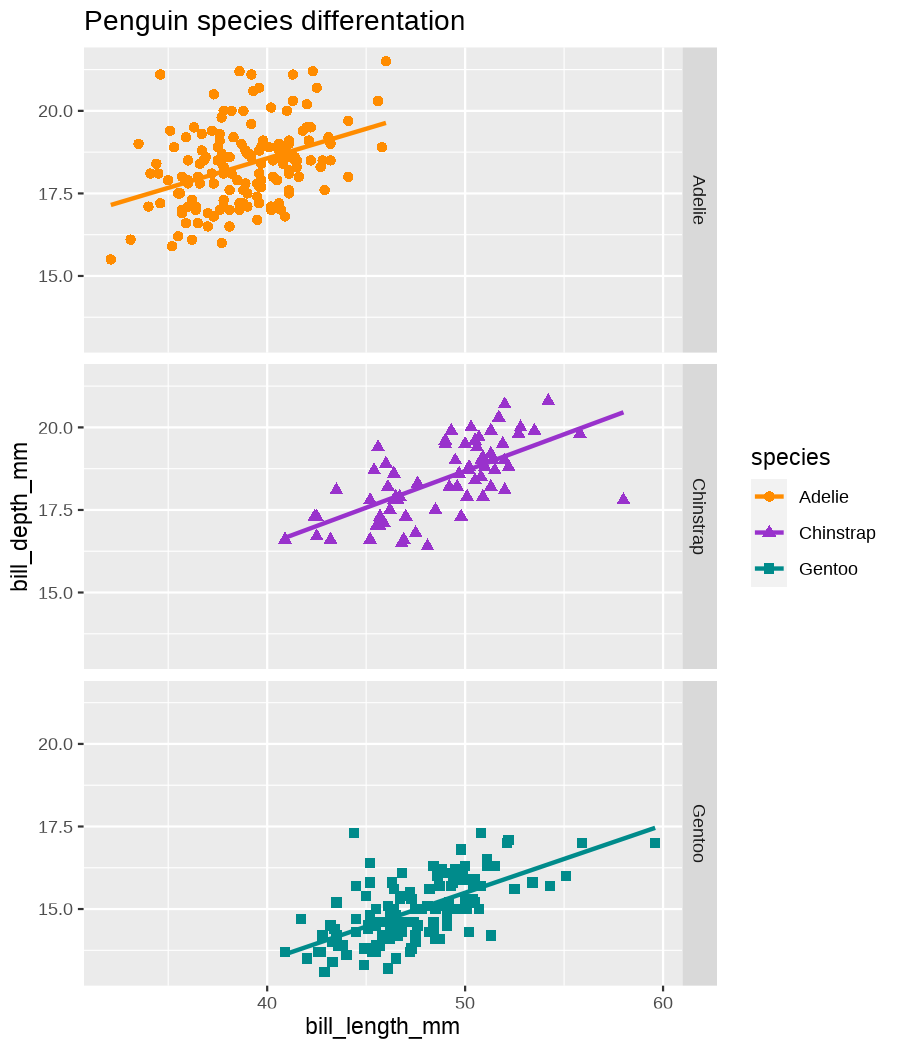<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + labs(title = "Penguin species differentation") + * labs(subtitle = * "based on bill depth and bill length") ``` ] .panel2-penguins_species3-auto[ 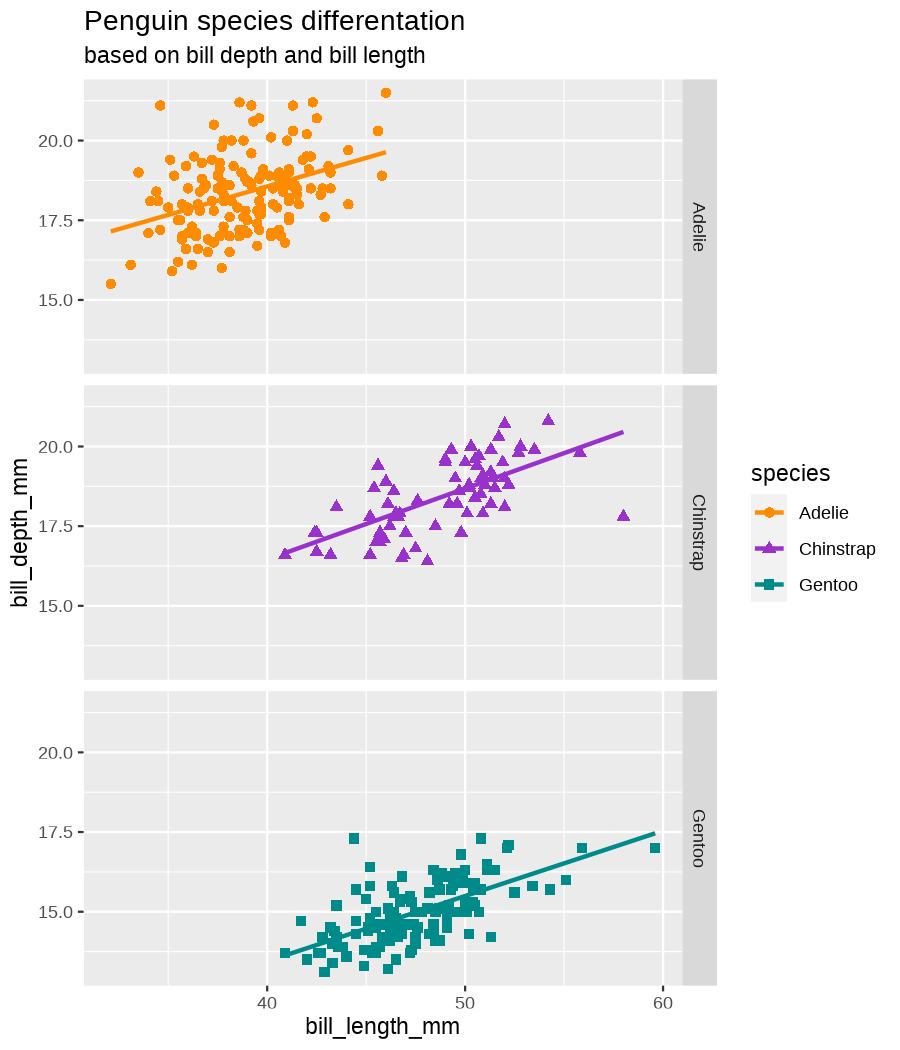<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + labs(title = "Penguin species differentation") + labs(subtitle = "based on bill depth and bill length") + * xlab("bill length (mm)") ``` ] .panel2-penguins_species3-auto[ 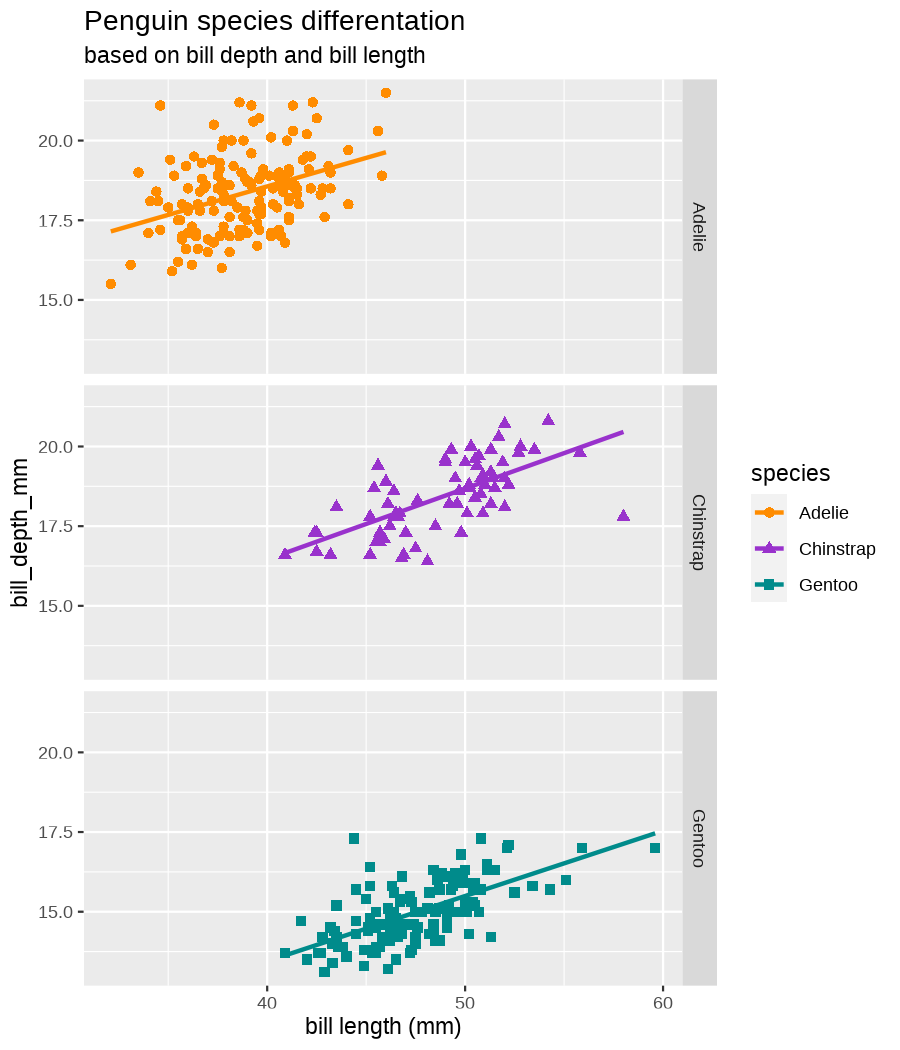<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + labs(title = "Penguin species differentation") + labs(subtitle = "based on bill depth and bill length") + xlab("bill length (mm)") + * ylab("bill depth (mm)") ``` ] .panel2-penguins_species3-auto[ 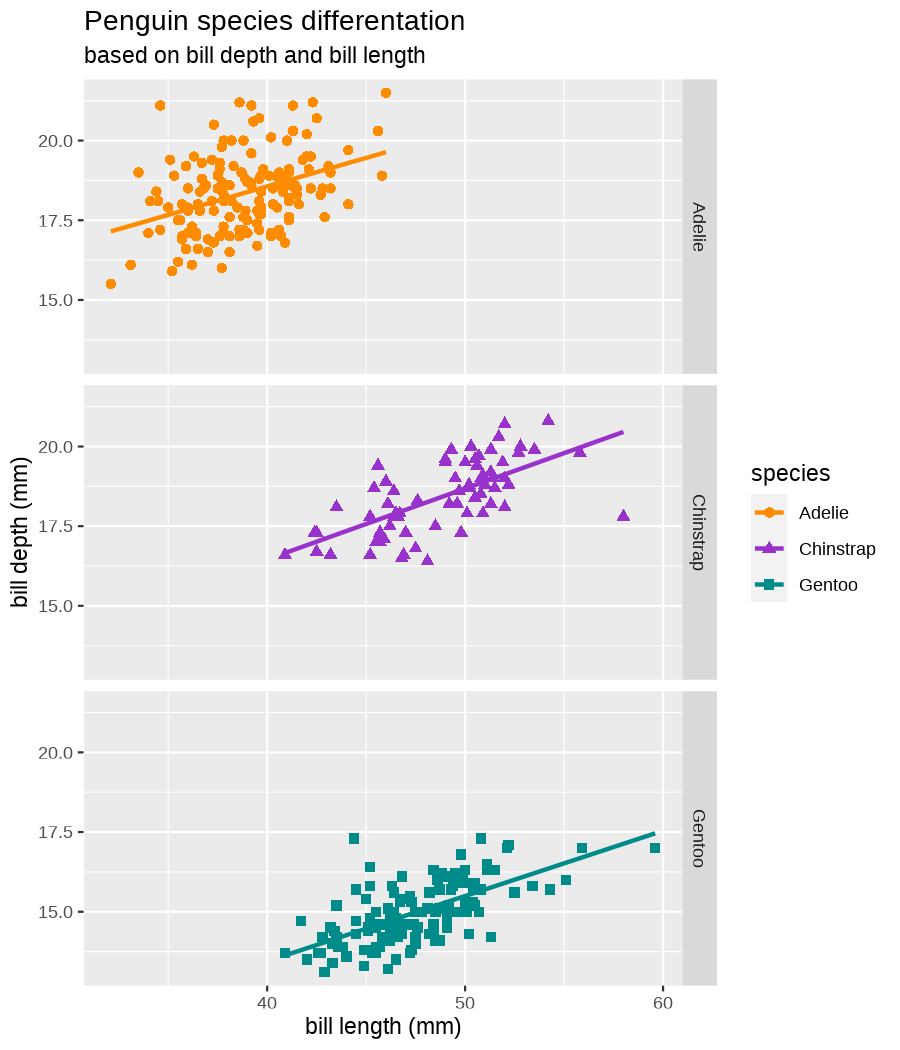<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + labs(title = "Penguin species differentation") + labs(subtitle = "based on bill depth and bill length") + xlab("bill length (mm)") + ylab("bill depth (mm)") + * theme_minimal() ``` ] .panel2-penguins_species3-auto[ 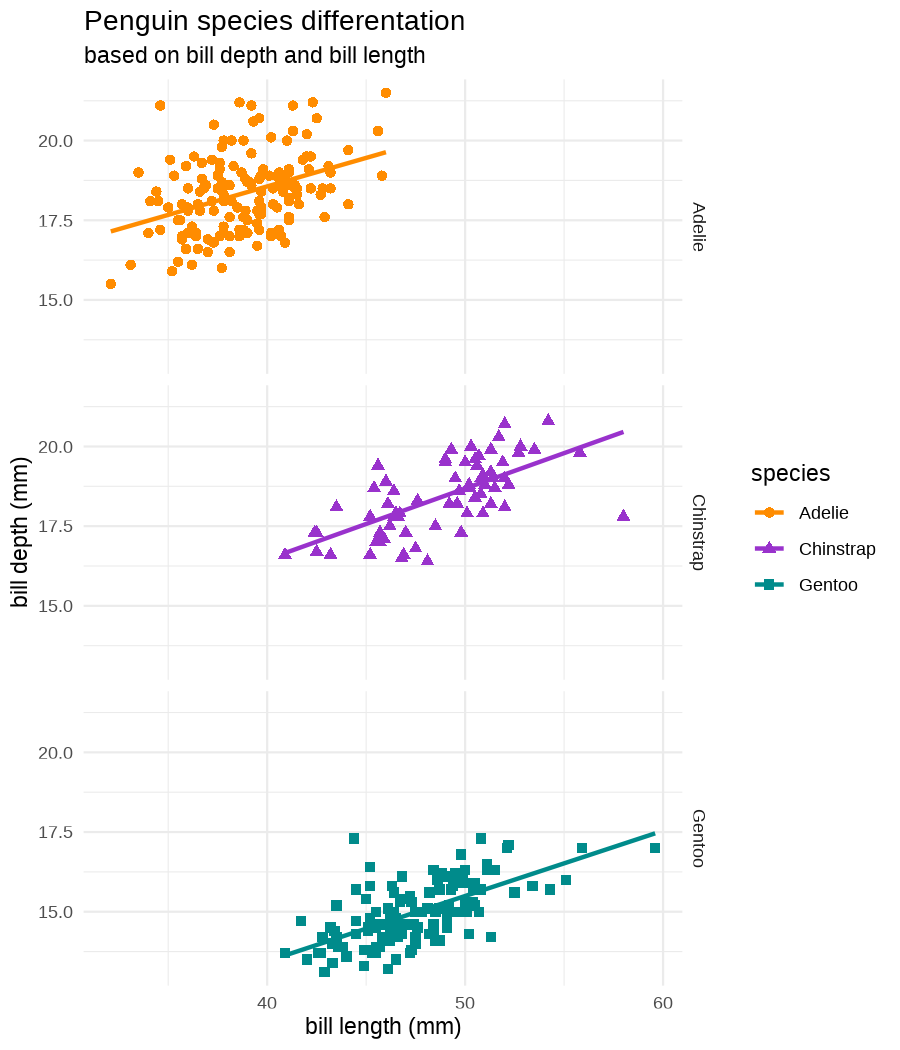<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + labs(title = "Penguin species differentation") + labs(subtitle = "based on bill depth and bill length") + xlab("bill length (mm)") + ylab("bill depth (mm)") + theme_minimal() + * theme( * plot.title = * element_text(face = "bold"), * plot.subtitle = * element_text(face = "italic")) ``` ] .panel2-penguins_species3-auto[ 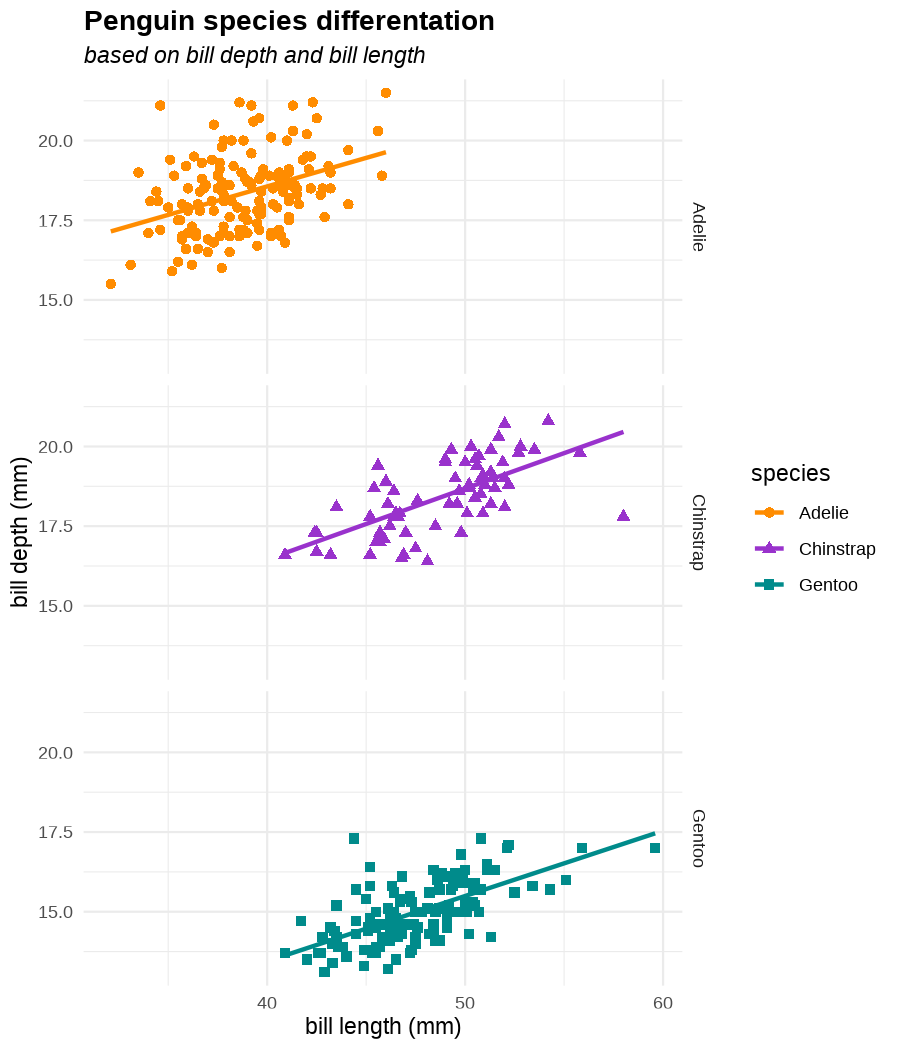<!-- --> ] --- count: false .panel1-penguins_species3-auto[ ```r penguins_facet + scale_color_manual( values = c("darkorange", "darkorchid", "cyan4") ) + labs(title = "Penguin species differentation") + labs(subtitle = "based on bill depth and bill length") + xlab("bill length (mm)") + ylab("bill depth (mm)") + theme_minimal() + theme( plot.title = element_text(face = "bold"), plot.subtitle = element_text(face = "italic")) + * theme(legend.position = "none") ``` ] .panel2-penguins_species3-auto[ 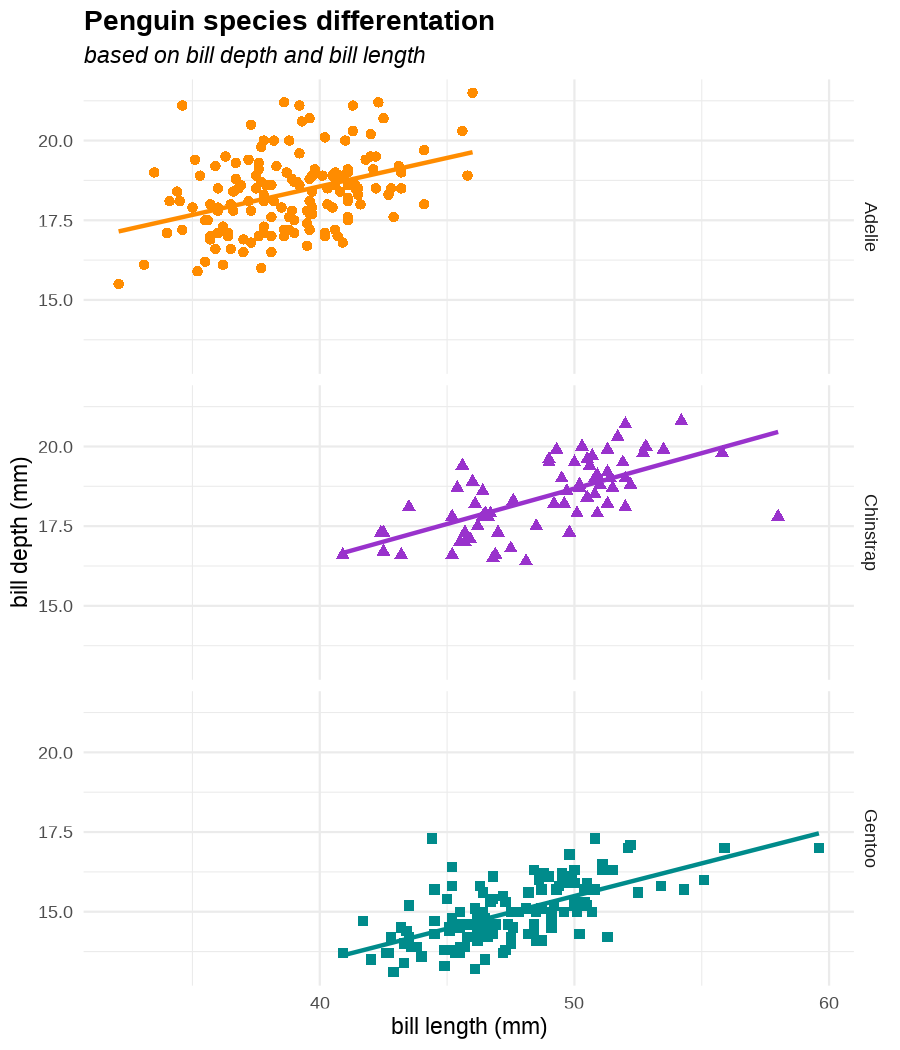<!-- --> ] <style> .panel1-penguins_species3-auto { color: black; width: 51.5789473684211%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel2-penguins_species3-auto { color: black; width: 46.421052631579%; hight: 32%; float: left; padding-left: 1%; font-size: 50% } .panel3-penguins_species3-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 50% } </style> ??? OK, that was the basic graph, but we can modify everything: - change of color, - split into sub-graphs, - adding graph title (and subtitle, caption) - adding axes titles - styling --- class: inverse, center, middle # <svg aria-hidden="true" role="img" viewBox="0 0 640 512" style="height:1em;width:1.25em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:currentColor;overflow:visible;position:relative;"><path d="M64 96c0-35.3 28.7-64 64-64H512c35.3 0 64 28.7 64 64V352H512V96H128V352H64V96zM0 403.2C0 392.6 8.6 384 19.2 384H620.8c10.6 0 19.2 8.6 19.2 19.2c0 42.4-34.4 76.8-76.8 76.8H76.8C34.4 480 0 445.6 0 403.2zM281 209l-31 31 31 31c9.4 9.4 9.4 24.6 0 33.9s-24.6 9.4-33.9 0l-48-48c-9.4-9.4-9.4-24.6 0-33.9l48-48c9.4-9.4 24.6-9.4 33.9 0s9.4 24.6 0 33.9zM393 175l48 48c9.4 9.4 9.4 24.6 0 33.9l-48 48c-9.4 9.4-24.6 9.4-33.9 0s-9.4-24.6 0-33.9l31-31-31-31c-9.4-9.4-9.4-24.6 0-33.9s24.6-9.4 33.9 0z"/></svg> Hands-on